Table 1. Crystallographic data and refinement statistics for KPN03535 (PDB code 3f1z).
Values in parentheses are for the highest resolution shell.
| Space group | P212121 |
| Unit-cell parameters (Å) | a = 97.42, b = 105.51, c = 181.25 |
| Data collection | |
| Wavelength (Å) | 0.9792 [Se peak (λ1)] |
| Resolution range (Å) | 29.9–2.46 (2.52–2.46) |
| No. of observations | 509996 |
| No. of unique reflections | 68362 |
| Completeness (%) | 99.8 (99.7) |
| Mean I/σ(I) | 15.4 (2.5) |
| Rmerge on I† (%) | 11.1 (69.6) |
| Model and refinement statistics | |
| Resolution range (Å) | 29.9–2.46 |
| No. of reflections (total) | 68310‡ |
| No. of reflections (test) | 3458 |
| Completeness (%) | 99.7 |
| Data set used in refinement | λ1 |
| Cutoff criterion | |F| > 0 |
| Rcryst§ | 0.192 |
| Rfree§ | 0.228 |
| Stereochemical parameters | |
| Restraints (r.m.s.d. observed) | |
| Bond angle (°) | 1.70 |
| Bond length (Å) | 0.017 |
| Average isotropic B value (Å2) | 38.2¶ |
| ESU†† based on Rfree (Å) | 0.22 |
| Protein residues/atoms | 1182/9162 |
| Water/PEG molecules | 323/2 |
R
merge = 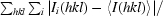
 .
.
Typically, the number of unique reflections used in refinement is slightly less than the total number that were integrated and scaled. Reflections are excluded owing to systematic absences, negative intensities and rounding errors in the resolution limits and unit-cell parameters.
R
cryst = 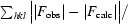
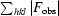 , where F
calc and F
obs are the calculated and observed structure-factor amplitudes, respectively. R
free is as for R
cryst, but for 5.1% of the total reflections chosen at random and omitted from refinement.
, where F
calc and F
obs are the calculated and observed structure-factor amplitudes, respectively. R
free is as for R
cryst, but for 5.1% of the total reflections chosen at random and omitted from refinement.
This value represents the total B that includes TLS and residual B components.
