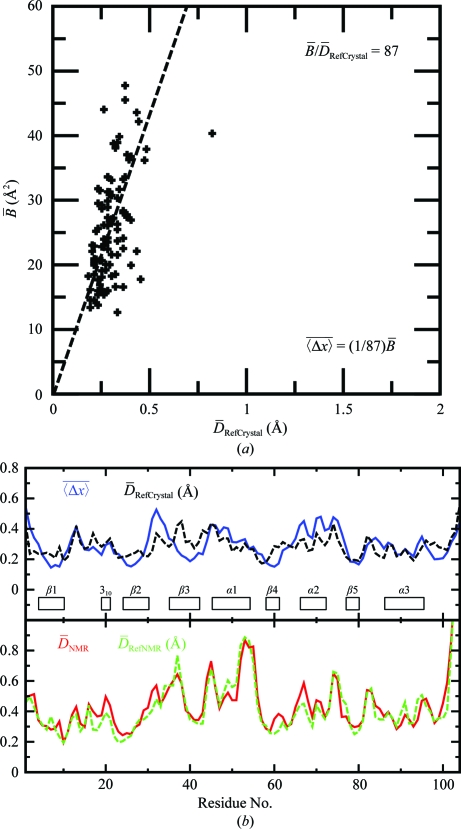
Figure 3.
Comparison of crystallographic B values and NMR displacements (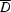 ). (a) Linear least-squares fit of the crystallographic
). (a) Linear least-squares fit of the crystallographic 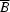 values (2)
versus the corresponding displacements
values (2)
versus the corresponding displacements 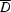 (1) in the reference crystal structure, yielding the c value in (3) for NP_247299.1, with c = 1/87. (b) Plots of the per-residue polypeptide backbone displacements versus the NP_247299.1 sequence. Upper panel, crystal structure and reference crystal structure. Lower panel, NMR structure and reference NMR structure. For the crystal structure, per-residue displacements were calculated from the
(1) in the reference crystal structure, yielding the c value in (3) for NP_247299.1, with c = 1/87. (b) Plots of the per-residue polypeptide backbone displacements versus the NP_247299.1 sequence. Upper panel, crystal structure and reference crystal structure. Lower panel, NMR structure and reference NMR structure. For the crystal structure, per-residue displacements were calculated from the 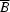 values with (2) and (3). For the NMR structure and the two reference structures, the data correspond to the per-residue displacements calculated with (1). The locations of regular secondary structures are indicated at the bottom of the upper panel.
values with (2) and (3). For the NMR structure and the two reference structures, the data correspond to the per-residue displacements calculated with (1). The locations of regular secondary structures are indicated at the bottom of the upper panel.