Abstract
Transport of protein and RNA cargoes between the nucleus and cytoplasm (nucleocytoplasmic transport) is vital for a variety of cellular functions. The studies of kinetics involved in such processes have been hindered by the lack of quantitative tools for measurement of the nuclear and cytosolic fractions of an intracellular protein at the single cell level for a cell population. In this report, we describe using a novel method, microfluidic electroporative flow cytometry, to study kinetics of nucleocytoplasmic transport of an important transcription factor NF-κB. With data collected from single cells, we quantitatively characterize the population-averaged kinetic parameters such as the rate constants and apparent activation barrier for NF-κB transport. Our data demonstrate that NF-κB nucleocytoplasmic transport fits first-order kinetics very well and is a fairly reversible process governed by equilibrium thermodynamics.
Introduction
A eukaryotic cell is divided by the nuclear membrane into two compartments with efficient and selective interchange of proteins, nucleic acids, and small molecules. Transport of protein and RNA cargoes between the nucleus and cytoplasm has been recognized as a critical process for the molecules to carry out their cellular functions 1–3. Nucleocytoplasmic transport is often regulated via upstream signal transduction pathways that lead to protein modifications or the masking or unmasking of the nuclear transport signals. As expected, alterations in any of the regulatory steps can result in the mislocalization of a protein leading to various disease states ranging to metabolic disorders to cancer 4. Not surprisingly, modification of the nucleocytoplasmic transport of specific target molecules has been proposed and practiced as an interesting therapeutic approach for disease treatment 4.
NF-κB is a family of dimeric transcription factors that regulate cellular stress responses, cell division, apoptosis, and inflammation and are prime examples of transcription factors that undergo nucleocytoplasmic transport in order to regulate transcription and gene expression 5–7. NF-κB activity is regulated by its cytoplasmic inhibitor IκB. IκB binds to NF-κB, masks its nuclear localization signal (NLS) and retains it in the cytoplasm preventing its association with DNA 8. Signals from extracellular stimuli (TNFα, IL-1, LPS and DNA-damaging agents etc.) are transduced via the IκB kinase (IKK) complex. Activated IKK phosphorylates IκB, which leads to its ubiquitination and proteolysis. This allows NF-κB to translocate to the nucleus, bind DNA and regulate gene expression. Predominant nuclear localization of NF-κB has been found in breast, ovary, colon, pancreas and thyroid tumor cells 9.
Traditionally, nucleocytoplasmic transport has been studied using fluorescence microscopy and/or subcellular fractionation followed by Western blotting 8, 10–12. Although capable of revealing protein transport in real time at the single cell level, fluorescence microscopy is limited by the number of cells that can be interrogated due to the small frame size. Thus, the data generated by fluorescence imaging may not provide an accurate description of an entire cell population. In comparison, subcellular-fractionation based analyses require homogenization of the cells such that the data reflect only the average properties of the population without single cell resolution. Neither approach is ideal for quantitative studies of the population behavior with single cell resolution. Single-cell tools such as laser scanning cytometry (LSC) 13, 14 or imaging flow cytometry15, 16 based on array detectors (e.g. CCD cameras) permit identification of the subcellular localization by conducting automated image analysis of solid-phase or flowing samples. Unfortunately, the image analysis algorithm is complex and lacks robustness and consistency for quantitative measurements. More importantly, the throughput of these techniques is ultimately limited by the exposure time needed to form images with sufficient spatial resolution and the response time/sensitivity of the array detectors.
In this study, we use a single-cell technique that combines electroporation with flow cytometry, referred to as Electroporative Flow Cytometry (EFC)17, for obtaining quantitative information on the kinetics of nucleocytoplasmic transport. Our approach does not require obtaining images of cells and therefore permits the use of a single element detector (e.g. a photomultiplier tube), which is essential for high throughput and easy quantification. Taking advantage of the fact that the release of an intracellular protein due to electroporation is dependent on the protein’s subcellular location, we quantify the cytosolic and nuclear fractions of NF-κB at the single cell level for a cell population during its nucleocytoplasmic transport. The data allow us to calculate the rate constants and the apparent activation barrier by fitting the process with first-order kinetics. The results indicate that the nucleocytoplasmic transport of NF-κB is governed by equilibrium thermodynamics and the transport process is more reversible than the prototypical nuclear import of proteins with NLS.
Experimental
Microchip fabrication
Microfluidic electroporative flow cytometry (EFC) devices were fabricated based on PDMS using the standard soft lithography method described previously 17, 18. The microscale patterns were first created using a computer-aided design software (FreeHand MX) and then printed out on high-resolution (5080 dpi) transparencies. The transparencies were used as photomasks in photolithography on a negative photoresist (SU-8 2010, MicroChem). The thickness of the photoresist and hence the depth of the channels was ~25 μm. The pattern of channels in the photomask was replicated in SU-8 after exposure and development. The microfluidic channels were molded by casting a layer (~5 mm) of PDMS prepolymer mixture (General Electric Silicones RTV 615, MG chemicals) with a mass ratio of A:B = 10:1 on the photoresist/silicon wafer master. The prepolymer mixture was cured at 85°C for 2 hrs in an oven and then peeled off from the master. Glass slides were cleaned in a basic solution (H2O: NH4OH (27%) : H2O2 (30%) = 5:1:1, volumetric ratio) at 75°C for 1 hr and then rinsed with DI water and blown dry. The surfaces of the PDMS chip and a glass slide were oxidized using a plasma cleaner (Harrick). The PDMS chip was then immediately brought into contact against the slide after oxidation to form closed channels.
Cell sample preparation
CHO/GFP-NFκBp65 cell line (Panomics) was created by co-transfection of an expression vector for a fusion protein of turboGFP (Evrogen) and human NFκBp65, as well as pHyg into Chinese hamster ovary (CHO) cells. CHO cells were subcultured for every 2 days in Hams F12Kmedia supplemented with 10% fetal bovine serum,100 IU/ml penicillin G, 100 μg/ml streptomycin and 100 μg/ml hygromycin. The harvested cells were either cultured in petri dishes or 96 well plates for 24 hr before starvation for 4 hr in serum free media. To stimulate adherent cells, the complete Hams F12Kmedia supplemented with IL-1 β at 20 ng/ml was used to replace starvation medium. Cells were detached by addition of 0.25% trypsin-EDTA for 1 min and washed twice by an electroporation buffer (1 mM MgSO4, 8 mM Na2HPO4, 2 mM KH2PO4, and 250 mM sucrose, pH 7.2). To stimulate cells in suspension, cells (1×106/ml) were suspended in the electroporation buffer with 20 ng/ml IL-1 β and incubated at the indicated temperature for the designated periods of time. All cell samples were centrifuged at 300 × g for 10 min and resuspended in the electroporation buffer at a final density of 1×107 cells /ml before a microfluidic EFC experiment.
Fluorescence microscopy
The microfluidic device was mounted on an inverted fluorescence microscope (IX-71, Olympus, Melville, NY) with a 40X dry objective (NA= 0.60). The epifluorescence excitation was provided by a 100W mercury lamp, together with brightfield illumination. The excitation and emission from CHO cells were filtered by a fluorescence filter cube (exciter HQ480/40, emitter HQ535/50, and beamsplitter Q505lp, Chroma Technology). To observe cells after electroporation, cells were transferred from the microchip reservoir to a 96 well plate and then centrifuged for 10 min at 300 × g to settle the cells to the bottom before imaging. The real-time observation and imaging of NFκB-GFP translocation in suspension cells were conducted by placing cells on a poly-L-lysine coated glass slide.
Microchip operation and cytometry detection
The microfluidic chip was mounted on the microscope. To prevent intracellular proteins from sticking to channel walls, the microfluidic channel was pre-treated by a 10% pluronic F68 solution (Sigma) for 1 hr followed by electroporation buffer for 15 min prior to all experiments. The 3 inlets of the channel were connected to a syringe pump (PHD infusion pump, Harvard Apparatus) through plastic tubing. The volumetric flow rates were set at 2 μl/min for the sample channel inlet and 10 μl/min for each of two side channel inlets. With a cell density of 1×107 cells/ml, 250–300 cells flowed through the laser detection spot per second. To prevent released proteins from interfering with the fluorescence signal of cells, the anode was placed in one of the side inlets while the cathode was located in the sample inlet so that the proteins went into the side inlet (Fig. 1a). A high voltage power supply (PS350, Stanford Research Systems) was used to generate a constant direct current (DC) voltage between the electrodes. A 488 nm laser beam (air-cooled 100 mW argon ion laser, Spectra-Physics) was focused in the microfluidic channel at 150 μm downstream of the intersection (Fig. 1a). The laser beam was spectrally filtered by a 10LF10-488 bandpass filter (Newport) before its intensity was adjusted by neutral density filters (Newport). The laser was introduced into the microscope through laser port B (Olympus) and a fluorescence filter cube (505DCLP dichroic beamsplitter, D535/40 emission filter, Chroma Technology) before it was finally focused by the objective into the microfluidic channel. The emission light was collected by the same objective and converted into current by a photomultiplier tube (R9220, Hamamatsu) biased at 500 V. The photocurrent was amplified by a low noise current preamplifier (SR570, Standard Research System) with the cutoff frequency and sensitivity set at 30K Hz and 20 μA/V, respectively. Cell autofluorescence generated only negligible signals with these optimized parameters. The current was then converted to voltage and input into a PCI data acquisition card (PCI-6254, National Instruments) operated by LabView software (National Instruments). The data were processed by programs written in MATLAB to extract histograms of the fluorescence for a cell population. The voltage signal ranging from 1 mV to 1 V was converted to 4 decade logarithmic voltage scale and then 256 scale channels, due to the relative small sample size of 6000–10000 cells in each histogram.
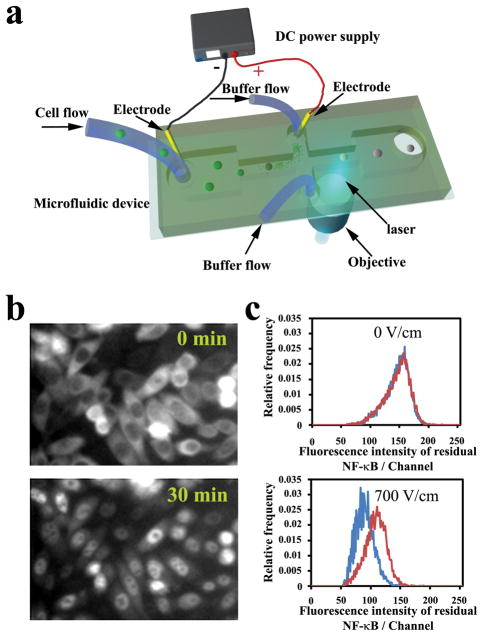
Figure 1.
(a) Schematic of the microfluidic electroporative flow cytometry device. The microfluidic device is mounted on a microscope that allows incident laser from a 40X objective to focus in the channel. A DC power supply generates a constant high electric field in the narrow sections to electroporate cells and release intracellular proteins. The width of the wide sections of the horizontal channel is 400 μm and that of the narrow section is 40 μm. The channel depth is 25 μm. (b) Fluorescence images of CHO cells expressing GFP labeled NF-κB. The cells were stimulated by 20 ng/ml IL-1 β at 37 °C at time 0. (c) Comparison of histograms of residual NF-κB between stimulated cells (red) and cells without stimulation (blue) under electric fields of 0 and 700 V/cm. The electroporation duration was 120 ms. The stimulation was conducted at 37 °C for 30 min.
Conversion of fluorescence signal into MESF
The quantification of fluorescence intensity from single cells using Molecules of Equivalent Soluble Fluorophores (MESF) units was described previously 17, 19, 20. In brief, a working curve was first established by screening beads with different predefined MESF values (high level Quantum FITC MESF Kits, Bangs Laborotories) using our microfluidic EFC devices. The peak channel number of the beads was plotted against their known MESF values (ESI Fig. S2). Using the working curve, the fluorescence intensity in channel numbers was converted into MESF. The percent of nuclear NF-κB for a cell population was calculated by using the MESF system and considering the average MESF value of CHO/GFP-NF-κBp65 without electroporation to be 100%.
Western blot analysis of nucleocytoplasmic transport
Cells were washed with 1X PBS supplemented with 200 μg/ml STI (soybean trypsin inhibitor) before they were suspended in 400 μl cold buffer A (10 mM Hepes, pH 7.9, 10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA, 1 mM DTT, 0.5 mM PMSF, 1 mM NaOV, 2X protease inhibitor cocktail) and allowed to swell on ice for 10 min. NP-40 was added to cells to achieve a final concentration of 0.2%. The homogenates were incubated on ice for another 5 min and then centrifuged at 20,000 × g for 30 sec. The supernatants were saved as cytosolic fractions while the nuclear pellets were washed with 200 μl detergent-free buffer A. The nuclear fractions were then resuspended in 300 μl SDS-sample buffer (25% sucrose, 2.5% SDS, 25 mM Tris/2.5 mM EDTA, 0.25 % pyronin Y) by using a 1-ml syringe with a 22G1/2 needle to shear the chromosomal DNA. The resulting samples were then subjected to SDS-PAGE and Western blot analysis with an anti-RelA antibody (Santa Cruz Biotechnology).
Results and Discussion
We used a Chinese hamster ovary (CHO) cell line expressing NF-κB with its p65 component fused to green fluorescent protein (Panomics). An improved version of microfluidic electroporative flow cytometry was used in this study 17. As shown in Fig. 1a, cells were allowed to flow through a microfluidic channel with a narrow middle section under hydrodynamic focusing. A constant voltage was established across two reservoirs of the microfluidic channels such that only the field intensity in the narrow section was high enough to produce electroporation 18. The nanoscale pores generated in the plasma membrane of cells by electroporation allowed intracellular molecules to be released into the surrounding solution 21–26. As an improvement to the previous design 17, in the current system the released proteins (including GFP labeled NF-κB) moved into the upper reservoir under the influence of the electric field without interfering with the laser-induced fluorescence detection while the remaining single cells (after release) flowed to the downstream laser detection point and generated signals that were collected by a photomultiplier tube (PMT). It is worth noting that given the intensive conditions of electroporation used in EFC, cells under examination lose their viability and biological functions after the procedure.
We found that the cytosolic fraction of a protein was released more readily during electroporation than the nuclear fraction of the same protein, presumably due to lack of an enclosure provided by the nuclear envelope and closer proximity to the plasma membrane. As shown in Fig. 1b, when NF-κB was activated (induced by IL-1β), a substantial portion moved into the nucleus. Consistent with this, the stimulated cell population retained a significantly larger amount of residual NF-κB after electroporation than did the unstimulated population. Such a difference could readily be observed from the histograms generated by the two populations (Fig. 1c, lower panel) and fluorescence images (ESI Fig. S1). In comparison, when there was no electric field, the device worked similarly to a conventional flow cytometer and no difference between the populations was detected (Fig. 1c, upper panel). Such a relationship between the electroporation-based release and the subcellular localization of the protein serves as the basis for our detection of nucleocytoplasmic transport without imaging.
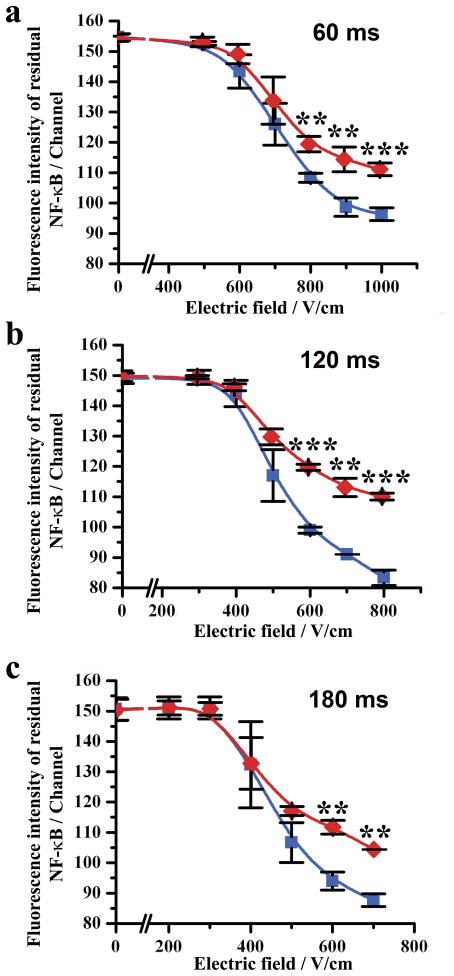
The operational parameters of electroporation such as the field intensity and duration affected how significant the difference between the cell population with translocation and the one without translocation was. As shown in Fig. 2, in general, the difference in the residual NF-κB amount between the two populations became larger when the field intensity increased. However, the difference did not increase monotonically with the field duration. We found that the difference was the largest when the field duration was 120 ms (compared to 60 and 180 ms) (Fig. 2b). Based on this optimization, we determined that a set of electric parameters (120 ms and 700 V/cm) were suitable as the operating conditions for EFC screening of this particular protein/cell system (800 V/cm was avoided due to the proneness to cell clogging in a very high field).
Figure 2.
The mean fluorescence intensity of a cell population generated by residual NF-κB after flow-through electroporation under different field intensities. Cell populations stimulated by IL-1β (red) and the ones without stimulation (blue) are compared when the field duration is (a) 60, (b) 120 and (c) 180 ms. The experiments were replicated in triplets to generate error bars. The cell stimulation was conducted by incubating with 20 ng/ml IL-1β for 0.5 h at 37°C. The difference between the two populations is considered statistically significant when P value is less than 0.01 (**) and 0.001 (***).
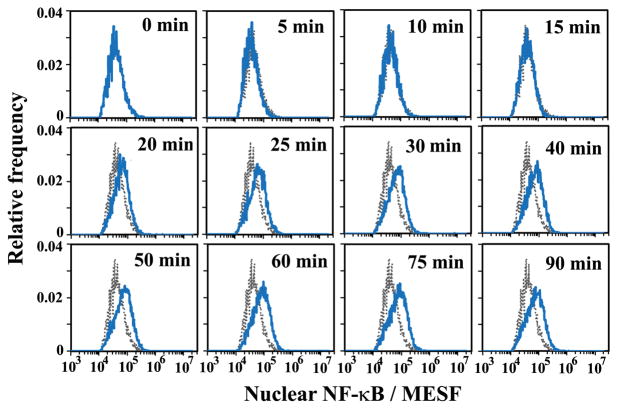
By quantifying the release of two marker intracellular proteins (p38, which is located in the cytosol and Sp1, which is nuclear) 11 by electroporation using Western blot analysis 27, we also estimated that, with an electroporation field intensity >600 V/cm and duration >100 ms, 100% of cytosolic proteins and 27% of nuclear proteins were released into the solution. It is worth noting that this estimation omits the effects of the type and amount of a protein on its release. Using these estimations and EFC data taken under the optimized operating conditions (120 ms and 700 V/cm), we were able to calculate the amount of NF-κB that was originally in the nucleus based on the residual NF-κB amount after electroporation at the single cell level (In this case the original amount was roughly a factor of 1.37 more than the residual amount, if we ignored the difference in the release pattern among cells). The fluorescence signal detected by the PMT was converted into Molecules of Equivalent Soluble Fluorophores (MESF) units for quantification purpose (ESI Fig. S2). Fig. 3 shows quantitatively that the histograms of nuclear NF-κB amount (in MESF) shifted toward the higher end over time after stimulation by IL-1β at 30 °C, indicating the gradual transport of NF-κB into the nucleus. These histograms present details about the cell populations during the transport event with single cell resolution.
Figure 3.
The histograms of nuclear NF-κB amount (in MESF) at different times after stimulation. The grey histogram was taken at time 0 and serves as a reference for the other histograms taken at later time points. The electroporative flow cytometry was conducted with an electroporation field of 700 V/cm and a duration of 120 ms. The cells were stimulated by 20 ng/ml IL-1β at 30 °C.
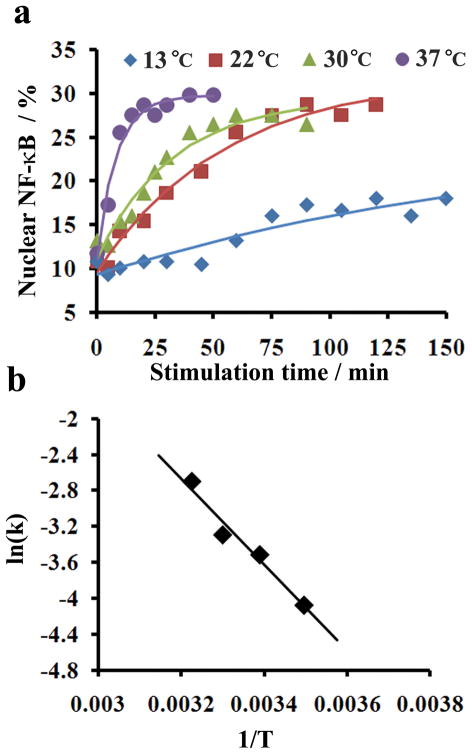
Taking advantage of the quantitative nature of our cytometry data, we were able to quantify the kinetics and activation barrier of the nucleocytoplasmic transport of NF-κB. The histograms such as those taken in Fig. 3 allow us to follow the progress of nucleocytoplasmic transport at the population level. We were able to generate plots indicating the change in the percent of nuclear NF-κB for the population over time at different stimulation temperatures between 13 and 37 °C (Fig. 4a) (Similar temperature range was used in a previous investigation28). In general, the transport occurred faster when the temperature was higher (Fig. 4a), consistent with what was revealed by Western blot analysis (ESI Fig. S3). The time course data in Fig. 4a fit very well (R2~0.94, Table 1) with a first-order process
Figure 4.
Kinetics of NF-κB nucleocytoplasmic transport. (a) Effect of temperature on nucleocytoplasmic transport rate. The data points are experimental data and the curves are fitted assuming first-order kinetics. (b) Arrhenius plot for calculating the apparent activation barrier Ea. Ea is calculated based on Arrhenius equation where k is the rate constant, R is the gas constant and C is a constant.
Table 1.
Kinetic parameters generated by fitting the data taken at different temperatures in Fig. 4a using Eqn.1. Nss is the steady state value of percentile nuclear NF-κB (the endpoint of the nucleocytoplasmic transport), N0 is the initial percentage of nuclear NF- κB at time 0, and k is the first-order rate constant.
| Temperature (K) | N0 (%) | Nss (%) | k (1/s) | R2 |
|---|---|---|---|---|
| 286 | 9.25 | 26.26 | 0.0050 | 0.8683 |
| 295 | 9.55 | 31.96 | 0.0182 | 0.9758 |
| 303 | 11.13 | 29.51 | 0.0303 | 0.9518 |
| 310 | 11.07 | 29.76 | 0.1189 | 0.9678 |
| (Eqn. 1) |
where N is the percent of nuclear NF-κB, Nss is the steady state value of the percent of nuclear NF-κB (the endpoint of the nucleocytoplasmic transport), N0 is the initial percent of nuclear NF-κB at time 0, and k is the first-order rate constant. The fitting of these curves generated values for k, N0, and Nss at various temperatures (Table 1). The percentage of nuclear NF-κB at the beginning of the translocation (N0 = 10.3±1.0%) and that at the end of the translocation (Nss = 29.2±2.3%) appear to be fairly consistent under different temperatures. In Fig. 4b, the Arrhenius plot generates an apparent activation barrier of 39.8 KJ/mol for the temperature range of 13–37 °C.
Interestingly, the apparent activation energy for the nucleocytoplasmic transport of NF-κB is lower than that measured for NLS-GFP (~43 kD) at 20–35 °C (~46–50 KJ/mol) 28. This reflects the fact that NF-κB translocation is more reversible than that of NLS-GFP. The phosphorylation of IκB leads to unmasking of NF-κB NLS and the nucleocytoplasmic transport. However, IκB is also a transcriptional target for NF-κB. Newly synthesized free IκB binds to nuclear NF-κB and exports the complex back to the cytoplasm to form the reverse process 8, 10. The fact that the equilibrium percent NF-κB (after translocation) in the nucleus is not strongly affected by the temperature (at 13–37 °C) not only suggests that the process is mostly governed by equilibrium thermodynamics, but also indicates that the overall process is not associated with significant enthalpy change (ΔH°) based on Van’t Hoff equation ( , where Keq is the equilibrium constant and R is the gas constant.)
Our approach generates highly quantitative kinetics data on the protein transport at the ensemble level by putting together information collected from single cells. These data provide the basis to compare the effectiveness of the machinery required for different molecules undergoing nucleocytoplasmic transport and to assess how these processes are affected by agents such as anticancer drugs. Such information is critical for understanding the hierarchical regulation of protein translocation and for discovering new drugs that block tumorigenesis by modifying the translocation of macromolecules.
Supplementary Material
Acknowledgments
We thank NSF CBET 1016547 and NIH NCI CA115465 for financial support of this research.
Footnotes
Electronic supplementary information (ESI) available: supplementary data.
References
- 1.Nigg EA. Nature. 1997;386:779–787. doi: 10.1038/386779a0. [DOI] [PubMed] [Google Scholar]
- 2.Nakielny S, Dreyfuss G. Cell. 1999;99:677–690. doi: 10.1016/s0092-8674(00)81666-9. [DOI] [PubMed] [Google Scholar]
- 3.Terry LJ, Shows EB, Wente SR. Science. 2007;318:1412–1416. doi: 10.1126/science.1142204. [DOI] [PubMed] [Google Scholar]
- 4.Kau TR, Way JC, Silver PA. Nat Rev Cancer. 2004;4:106–117. doi: 10.1038/nrc1274. [DOI] [PubMed] [Google Scholar]
- 5.Hoffmann A, Baltimore D. Immunol Rev. 2006;210:171–186. doi: 10.1111/j.0105-2896.2006.00375.x. [DOI] [PubMed] [Google Scholar]
- 6.Ghosh S, May MJ, Kopp EB. Annu Rev Immunol. 1998;16:225–260. doi: 10.1146/annurev.immunol.16.1.225. [DOI] [PubMed] [Google Scholar]
- 7.Hoffmann A, Natoli G, Ghosh G. Oncogene. 2006;25:6706–6716. doi: 10.1038/sj.onc.1209933. [DOI] [PubMed] [Google Scholar]
- 8.Hoffmann A, Levchenko A, Scott ML, Baltimore D. Science. 2002;298:1241–1245. doi: 10.1126/science.1071914. [DOI] [PubMed] [Google Scholar]
- 9.Rayet B, Gelinas C. Oncogene. 1999;18:6938–6947. doi: 10.1038/sj.onc.1203221. [DOI] [PubMed] [Google Scholar]
- 10.Nelson DE, Ihekwaba AEC, Elliott M, Johnson JR, Gibney CA, Foreman BE, Nelson G, See V, Horton CA, Spiller DG, Edwards SW, McDowell HP, Unitt JF, Sullivan E, Grimley R, Benson N, Broomhead D, Kell DB, White MRH. Science. 2004;306:704–708. doi: 10.1126/science.1099962. [DOI] [PubMed] [Google Scholar]
- 11.Zhou F, Hu J, Ma H, Harrison ML, Geahlen RL. Mol Cell Biol. 2006;26:3478–3491. doi: 10.1128/MCB.26.9.3478-3491.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.James CD, Moorman MW, Carson BD, Branda CS, Lantz JW, Manginell RP, Martino A, Singh AK. Biomed Microdevices. 2009;11:693–700. doi: 10.1007/s10544-008-9281-5. [DOI] [PubMed] [Google Scholar]
- 13.Deptala A, Bedner E, Gorczyca W, Darzynkiewicz Z. Cytometry. 1998;33:376–382. doi: 10.1002/(sici)1097-0320(19981101)33:3<376::aid-cyto13>3.0.co;2-q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harnett MM. Nat Rev Immunol. 2007;7:897–904. doi: 10.1038/nri2188. [DOI] [PubMed] [Google Scholar]
- 15.Arechiga AF, Bell BD, Solomon JC, Chu IH, Dubois CL, Hall BE, George TC, Coder DM, Walsh CM. J Immunol. 2005;175:7800–7804. doi: 10.4049/jimmunol.175.12.7800. [DOI] [PubMed] [Google Scholar]
- 16.Fanning SL, George TC, Feng D, Feldman SB, Megjugorac NJ, Izaguirre AG, Fitzgerald-Bocarsly P. J Immunol. 2006;177:5829–5839. doi: 10.4049/jimmunol.177.9.5829. [DOI] [PubMed] [Google Scholar]
- 17.Wang J, Bao N, Paris LL, Wang HY, Geahlen RL, Lu C. Anal Chem. 2008;80:1087–1093. doi: 10.1021/ac702065e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang HY, Lu C. Anal Chem. 2006;78:5158–5164. doi: 10.1021/ac060733n. [DOI] [PubMed] [Google Scholar]
- 19.Wang LL, Gaigalas AK, Abbasi F, Marti GE, Vogt RF, Schwartz A. J Res Natl Inst Standards Technol. 2002;107:339–353. doi: 10.6028/jres.107.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martin BR, Giepmans BNG, Adams SR, Tsien RY. Nat Biotechnol. 2005;23:1308–1314. doi: 10.1038/nbt1136. [DOI] [PubMed] [Google Scholar]
- 21.Ryttsen F, Farre C, Brennan C, Weber SG, Nolkrantz K, Jardemark K, Chiu DT, Orwar O. Biophys J. 2000;79:1993–2001. doi: 10.1016/S0006-3495(00)76447-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McClain MA, Culbertson CT, Jacobson SC, Allbritton NL, Sims CE, Ramsey JM. Anal Chem. 2003;75:5646–5655. doi: 10.1021/ac0346510. [DOI] [PubMed] [Google Scholar]
- 23.Lu H, Schmidt MA, Jensen KF. Lab Chip. 2005;5:23–29. doi: 10.1039/b406205a. [DOI] [PubMed] [Google Scholar]
- 24.Marc PJ, Sims CE, Bachman M, Li GP, Allbritton NL. Lab Chip. 2008;8:710–716. doi: 10.1039/b719301g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Valero A, Post JN, van Nieuwkasteele JW, Ter Braak PM, Kruijer W, van den Berg A. Lab Chip. 2008;8:62–67. doi: 10.1039/b713420g. [DOI] [PubMed] [Google Scholar]
- 26.Agarwal A, Wang MY, Olofsson J, Orwar O, Weber SG. Anal Chem. 2009;81:8001–8008. doi: 10.1021/ac9010292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhan Y, Martin VA, Geahlen RL, Lu C. Lab Chip. 2010;10:2046–2048. doi: 10.1039/c005152g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shulga N, Roberts P, Gu ZY, Spitz L, Tabb MM, Nomura M, Goldfarb DS. J Cell Biol. 1996;135:329–339. doi: 10.1083/jcb.135.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.