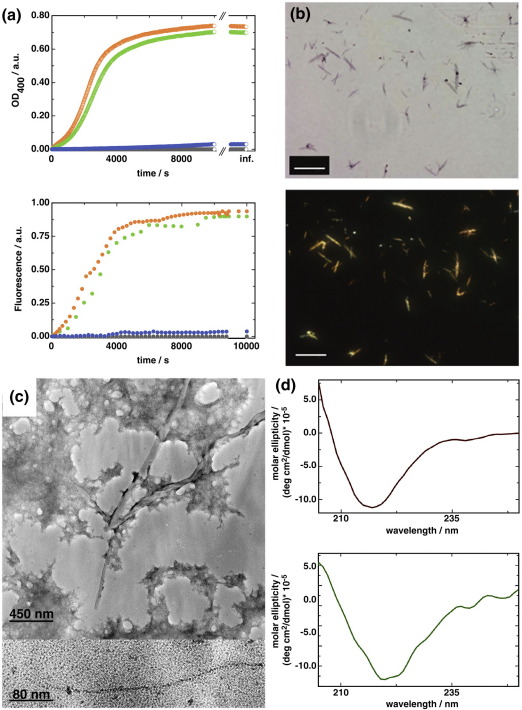
Fig. 2.
Amyloid formation by an Ig domain. (a) Monomeric CTLA4 was concentrated to 10 mg/ml and incubated for 24 h at room temperature. The solution was then applied to a size-exclusion chromatography column whereupon a stable dimeric species as well as higher molecular weight species and aggregates could be separated (see Supplementary Fig. S3a). Under the strong denaturing (but not reducing) conditions in electrospray ionization mass spectrometry only molecular weights that corresponded to the monomer were found (see Supplementary Fig. S3b, where a representative deconvoluted mass spectrum is shown). To further assay the aggregation, it was induced by incubating CTLA4 in phosphate-buffered saline (PBS) with 28% TFE and the scatter of light at 400 nm was followed in real time (see Refs. 10 and 20). Aggregate formation was followed by monitoring the scattering of 400 nm light (top) and Thioflavin T fluorescence increase (bottom) for monomer (orange), strand-swapped dimer (green), higher-order aggregate (blue) and disulfide-bonded maCTLA-4 (grey) upon incubation with TFE. Optical density (OD) and fluorescence are reported in arbitrary units (a.u.). For details of fluorescence measurements, see Supplementary Fig. S4. (b) Congo Red staining of amyloid fibrils. CTLA-4 amyloid fibrils (50 μl) were placed on a clean microscope slide and dried under air. A 10 μM solution of Congo Red in PBS at pH 7.4 was filtered several times through a 0.22-μm nucleopore filter and 50 μl was added onto the dried amyloid fibrils. A clean coverslip was placed onto the sample, which was then dried with a paper towel and sealed along the edges of the coverslip with nail varnish. The sample was imaged under normal and crossed-polarised light with a Nikon Eclipse TE2000U inverted microscope fitted with a charge-coupled device camera. Here we show aggregates imaged under bright-field conditions (top) and the same field of view with crossed-polarising filters introduced above and below the specimen (bottom). The scale bar indicates a length of 50 μm in both panels. (c) Electron microscopy of CTLA-4 aggregates. Aggregates were spun down in a benchtop centrifuge and stained with 1% uranyl acetate on carbon-coated electron microscopy copper grids. The samples were imaged on Kodak SO-163 film with a Tecnai F30 electron microscope (FEI) operating at 300 kV accelerating voltage. Here we show an electron micrograph of negatively stained aggregate; stranded structures can clearly be discerned. The close-up of an individual strand (bottom) shows the apparently bead-like arrangement of the Ig domains in the fibril. (d) Far-UV CD measurements were performed with a Chirascan spectrophotometer (Applied Photophysics) in a 1-mm path-length quartz cuvette at a concentration of 20 μM protein. Measurements were conducted in PBS (pH 7.5) for the CTLA4 before aggregate formation and after aggregate formation. The obtained ellipticities, θ, were converted to molar ellipticities, [θ], according to the equation , where c is the concentration of CTLA-4 in monomer units and l is the path length of the cuvette. A CD spectrum of monomeric ecCTLA4 is shown (top) and a CD spectrum of ecCTLA-4 after amyloid formation (bottom). The prominent minimum around 220 nm is indicative of β-strand secondary structural elements. See Supplementary Fig. S3 for assessment of protein aggregate formation and the absence of disulfide bonds.