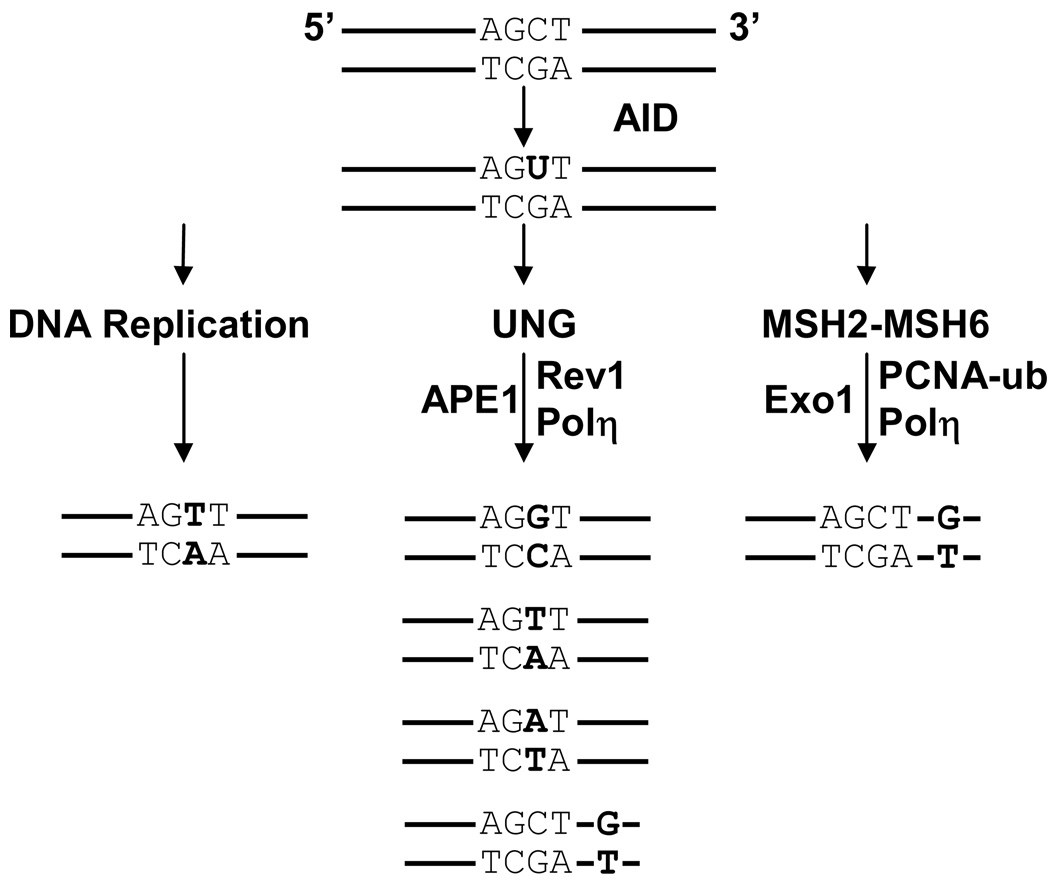
FIGURE 3.
Three error-prone pathways for processing U:G mismatches. AID deaminates dC to dU in a hotspot, WGCW. Mutations are shown in bold. (1) DNA replication. U mimics T, and replication from the 5’ strand will incorporate C:G to T:A transitions. (2) UNG recognition. UNG removes the U, and several possibilities could occur. Rev1 may bypass the abasic site with a C:G to G:C transversion. APE1 may nick the abasic site, and the 5’ strand would be extended by a low-fidelity polymerase to insert other transversions. Polη may extend the synthesis to insert G opposite a downstream template T, to cause A:T to G:C transitions. (3) MSH2-MSH6 recognition. The two proteins bind to the U:G mismatch and recruit Exo1 to form a gap. It is not known what triggers a nick for Exo1 to access the DNA. Mono-ubiquitinated PCNA brings in Polη to synthesize predominantly A:T mutations downstream of the initial deamination event.