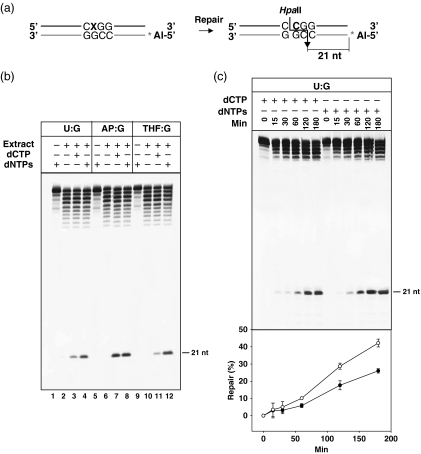
Figure 1.
Repair of base DNA damage by Arabidopsis cell extracts. (a) Schematic diagram of molecules used as DNA substrates. Double-stranded oligonucleotides contained a lesion (X = uracil or AP site) at an HpaII site on the upper strand. The Alexa Fluor-labeled 5′ end of the lower strand is indicated by a star. The size of the 5′ end-labeled fragment generated after HpaII digestion of the fully repaired product is indicated on the right. (b) DNA duplexes containing U, a natural AP site, or THF opposite G were incubated with Arabidopsis cell extract at 30°C for 3 h in a reaction mixture containing either dCTP or all four dNTPs, as indicated. Reaction products were digested with HpaII, separated in a 12% denaturing polyacrylamide gel and quantified by fluorescence scanning. (c) A DNA duplex containing a U:G mispair was incubated with Arabidopsis cell extracts at 30°C in a reaction mixture containing either dCTP or all four dNTPs. Reactions were stopped at the indicated times and products were analyzed as described above. The percentage of fully repaired DNA product for reactions containing only dCTP (•) or all four dNTPs (○) is shown in the bottom panel. Values are means with standard errors from two independent experiments.