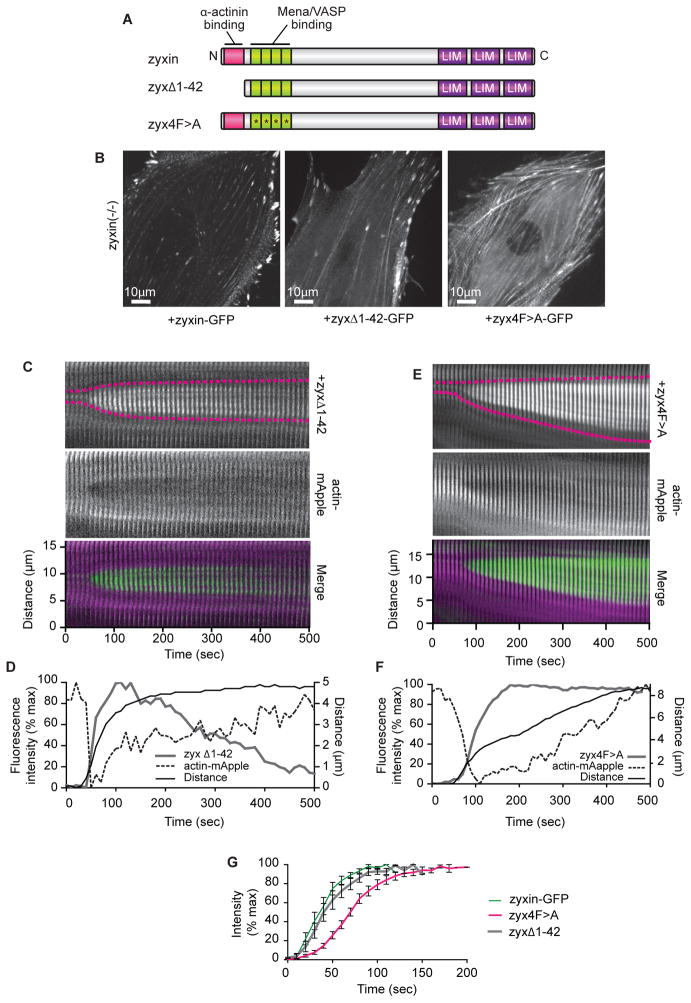
Figure 5.
Zyxin accumulation at SF strain sites is not dependent on zyxin’s interaction with α-actinin, but is facilitated by interaction with VASP.
(A) Schematic of wild-type zyxin and zyxin mutated to interfere with α-actinin binding (zyxΔ1-42), or Mena/VASP protein binding (zyx4F>A).
(B ) Mutant zyxin zyxΔ1-42 and zyx4F>A localize to SFs in zyxin (−/ −) cells.
(C) Kymograph of zyxΔ1-42 and actin-mApple expressed in zyxin (−/ −) cell. For merge, zyx 1-42 is green, actin is magenta.
(D) Intensity and distance graph showing zyxΔ1-42 accumulates at SF strain sites.
(E) Kymograph of zyx4F>A and actin-mApple expressed in zyxin (−/ −) cell. For merge, zyx4F>A is green, actin is magenta.
(F) Intensity and distance graph showing zyx4F>A accumulates at SF strain sites.
(G) Zyx4F>A (n=12) accumulation at stress fiber strain sites is slower than wild-type zyxin (n=13) (p<0.0001) or zyxΔ1-42 (n=12) (p=0.0002).
Data are shown as mean+/− SEM