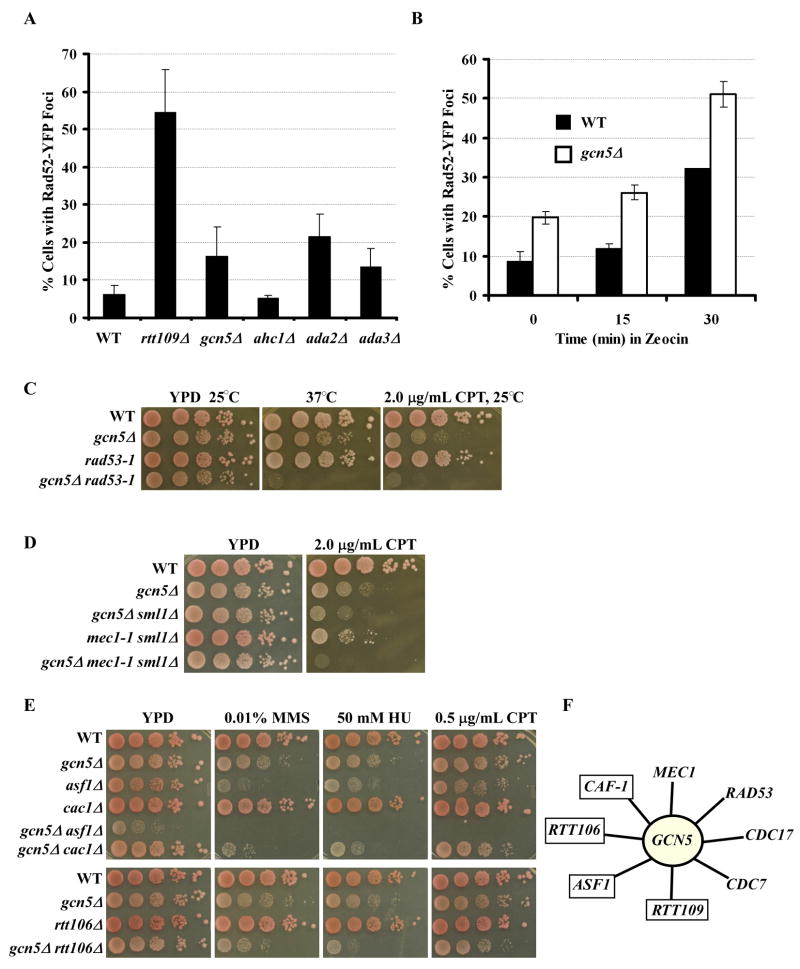
Figure 5.
GCN5 is involved in DNA replication-coupled nucleosome assembly. (A) Cells lacking GCN5, ADA2 or ADA3 exhibited a higher level of Rad52 foci compared to wild-type cells. The percentage of cells with Rad52-YFP foci was determined as described in Experimental Procedures. Data represent the mean percentage ± SEM. Mock experiments were also performed in which the genotype of each strain was unknown to the person who performed the experiment. (B) The gcn5Δ mutant cells exhibited higher levels of chromosome breaks than wild-type cells when challenged with a DNA damaging agent. Wild-type or gcn5Δ cells expressing Rad52-YFP were treated with zeocin for 0, 15, or 30 minutes and the percentage of cells containing Rad52 foci was determined. (C–D) The gcn5Δ mutation exhibited a synthetic genetic interaction with mutations in the checkpoint kinases RAD53 (C, rad53-1) and MEC1 (D, mec1-1). (E) The gcn5Δ mutant exhibited slow growth and increased DNA damage sensitivity with mutations in ASF1, CAC1, and RTT106. (F) A summary of genetic interactions observed for GCN5. Those in blue boxes indicate genes that are known to be involved in nucleosome assembly. Complete genetic data is presented in Figure S3.