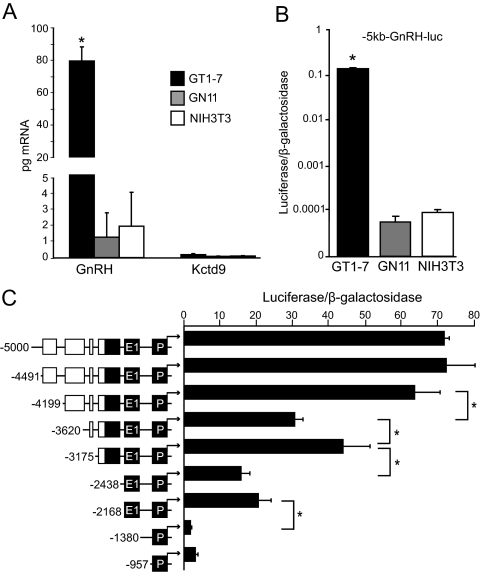
Figure 2.
The 5 kb of GnRH upstream sequence displays cell specificity similar to GnRH but not Kctd9 mRNA. A, RT-qPCR analysis of GnRH and Kctd9 mRNA from GT1-7, GN11, and NIH3T3 cells. Data are presented as picograms of mRNA of each gene normalized to cyclophilin B and indicate mean ± sd. Statistical analysis was performed using two-way ANOVA followed by post hoc least squares mean differences Tukey-Kramer HSD. *, Significantly different from GnRH mRNA levels in GN11 and NIH3T3 cells and Kctd9 mRNA levels in all three cell lines (P < 0.05). B, Transient transfection of the −5-kb GnRH gene sequence upstream of a luciferase reporter gene (−5 kb-GnRH-luc) into GT1-7, GN11, and NIH3T3 cells. Luciferase values were normalized to a cotransfected β-galactosidase reporter to control for transfection efficiency, and data were normalized to RSVe/RSVp-luc transfected in parallel. Data represent mean ± sd plotted on a logarithmic scale. Statistical analysis was performed using one-way ANOVA followed by post hoc Tukey Kramer HSD. *, Significantly different from GN11 and NIH3T3 cells (P < 0.05). C, Truncation analysis of −5 kb of the GnRH gene sequence in GT1-7 cells. Transient transfections were performed in GT1-7 cells using 5′ truncations of −5-kb-GnRH-luc reporter plasmids as indicated by schematic diagrams. Locations of truncations are denoted as base-pair positions upstream of the GnRH TSS to the left of schematic diagrams. Previously characterized GnRH regulatory elements are colored in black, and novel conserved sequence blocks are colored in white. Data represent luciferase/β-galactosidase values normalized to empty pGL3 vector and are shown as mean ± sd. *, Significant difference between constructs by one-way ANOVA and post hoc Tukey-Kramer HSD (P < 0.05). P, Promoter; E1, enhancer-1.