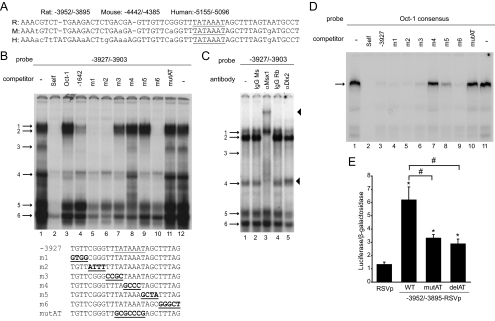
Figure 6.
The critical region of GnRH-E3 is regulated by Oct-1, Ms-1, and Dlx2. A, Rat, mouse, and human sequences corresponding to the GnRH-E3 critical region aligned by VISTA. Bases conserved in all three species are in capital letters. AT-rich region −3917/−3911 is underlined. B, EMSA was performed using GT1-7 nuclear extracts using a radiolabeled oligonucleotide probe containing −3927/−3903 of GnRH-E3 (lanes 1 and 12). Competition assays were performed with unlabeled oligonucleotides: −3927/−3903 (Self) (lane 2), Oct-1 consensus (Oct-1) (lane 3), Q50 homeodomain binding region in −1642/−1623 in GnRH-E1 (−1642) (lane 4), 4-bp scanning mutants of −3927/−3903 (m1–m6) (lanes 5–10), and mutation of AT-rich region −3917/−3911 (mutAT) (lane 11). Protein complexes are indicated by numbered arrows. Sequences of wild-type (WT) and mutant −3927/−3903 EMSA probes for competition assays are below the gel. The AT-rich −3917/−3911 region is underlined, and mutations are bold and underlined. C, EMSA with GT1-7 extracts using a radiolabeled probe containing −3927/−3903. Supershift assays were performed with mouse (Ms) and rabbit (Rb) IgG controls (lanes 2 and 4, respectively) and with antibodies recognizing Msx1 and Dlx2 (lanes 3 and 5, respectively). Protein complexes are indicated by numbered arrows, supershifted complexes are indicated by arrowheads. D, EMSA with GT1-7 nuclear extracts using a radiolabeled probe containing the Oct-1 consensus sequence. The Oct-1 complex is indicated by an arrow. Competition assays were performed with: Oct-1 consensus (Self) (lane 2), −3927/−3903 (−3927) (lane 3), scanning mutants m1–m6 in −3927/−3903 (lanes 4–9), and mutAT in −3927/−3903 (lane 10). E, Transient transfections were performed in GT1-7 cells using luciferase reporter plasmids containing the GnRH-E3 critical region (−3952/−3895) on RSVp containing a mutated (mutAT) or deleted (delAT)AT-rich region −3917/−3911. Data represent luciferase/β-galactosidase values normalized to empty vector and are shown as mean ± sd. *, Significantly different from RSVp; and #, significantly different from WT by one-way ANOVA and post hoc Tukey-Kramer HSD (P < 0.05).