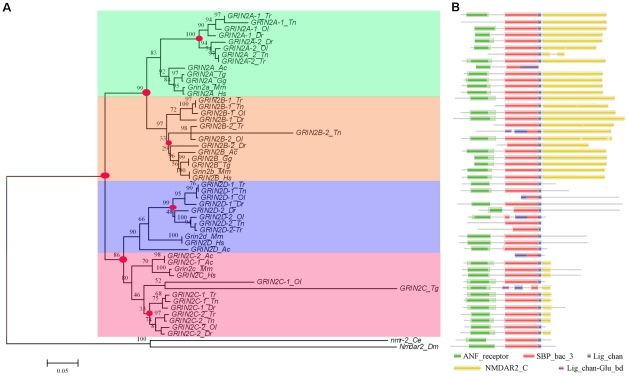
Figure 1. The phylogenetic tree (A) of NR2 proteins and their protein domain structure (B).
The bootstrap consensus phylogenetic tree was constructed with the fruitfly Nmdar2 and nematode nmr-2 protein as outgroup using the neighbor-joining method in MEGA4, and the numbers indicate the percentage bootstrap support. The evolutionary distances were computed using the Poisson correction method and are in the units of the number of amino acid substitutions per site. Red circles indicate the gene duplication event supported by the phylogenetic analysis. Dm, Drosophila melanogaster; Ce, Caenorhabditis elegans; Dr, Danio rerio; Tr, Takifugu rubripes; Tn, Tetraodon nigroviridis; Ol, Oryzias latipes; Ac, Anolis carolinensis; Gg, Gallus gallus; Tg, Taeniopygia guttata; Mm, Mus musculus; Hs, Homo sapiens. Protein domains are shown as boxes based on identification by Pfam.