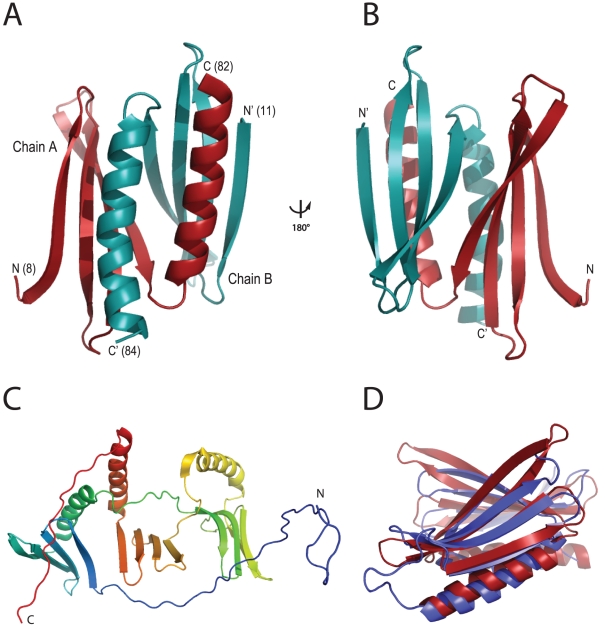
Figure 3. Ribbon backbone models for Pur-α proteins.
(A) Crystal structure of B. burgdorferi Pur-α with one monomer shown in red, the other in cyan. N- and C-termini are indicated with “N” and “C” respectively, followed by corresponding amino-acid positions in parentheses. (B) Identical to (A), with the structural model rotated 180° around the vertical axis. (C) Computational model for D. melanogaster Pur-α calculated with the program HHpred. Rainbow-color coding follows the peptide chain from N-terminus (blue) to C-terminus (red). It shows the secondary structure of the PUR repeats, but lacks information about the correct tertiary structure. (D) Superposition of the crystal structures of B. burgdorferi Pur-α (red, PDB-ID 3N8B) and D. melanogaster Pur-α repeats I-II (blue, PDB-ID 3K44) [9]. RMSD for Cα-carbon atoms is 2.1 Å. B. burgdorferi Pur-α forms an inter-molecular dimer, whereas PUR repeat I and PUR repeat II in D. melanogaster Pur-α form an intra-molecular dimer.