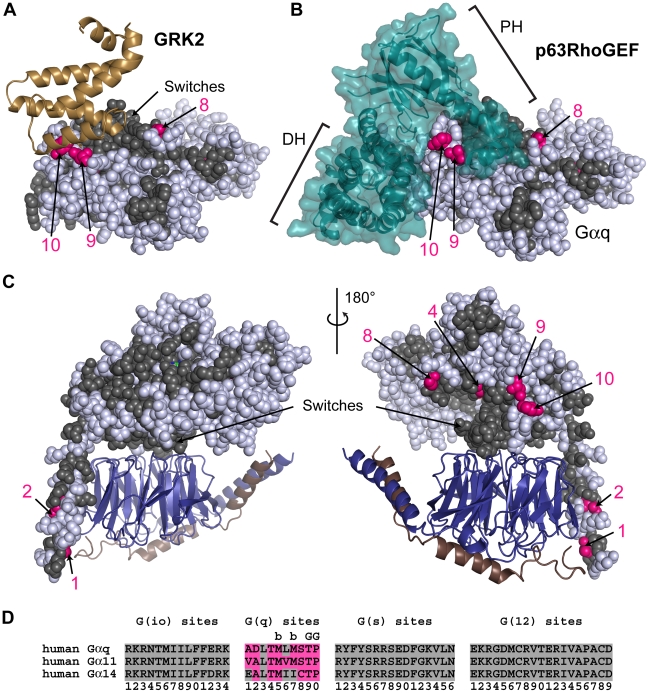
Figure 4. G(q) class-distinctive sites in structural context.
(A) The RH domain of GRK2, shown as a sand colored cartoon display, in complex with activated Gαi/q •GDP•Mg2+ •AlF4 − (PDB ID 2BCJ). In all structural panels in this figure, Gαi/q is shown as spheres with core residues colored gray if the residues are conserved between Gα subunits (either INV (invariant) or η (non-distinct) amino acids) while G(q)-distinctive sites are colored hot pink only if they contain a ∂ (distinct) amino acid. Non-core residues and d sites are colored white. G(q)-distinctive sites are numbered according to their position in the signature sequence (see panel (D)). (B) The DH and PH domains of p63RhoGEF, in a teal colored cartoon and surface display, binds to activated Gαi/q •GDP•Mg2+ •AlF4 − (PDB ID 2RGN). Gαi/q is in the same orientation as panel A. (C) Homology model of Gαq •GDP (sphere display) bound to Gβ•Gγ (deepblue/copper cartoon) heterodimer. Two orientations related by a 180° rotation about the vertical axis are shown. (D) Signature sequences are formed by grouping all distinctive sites for a given class together, removing all residues between individual distinctive sites of the noted class. The distinctive sites for each class are presented in order from the N-terminus to the C-terminus and numbered accordingly. Amino acids that correspond to the ∂ values at the G(q) site are colored hot pink. Sites that interact with GRK2 are denoted by ‘G’ above the site, while sites that are buried and not visible are denoted by ‘b’ above the site.