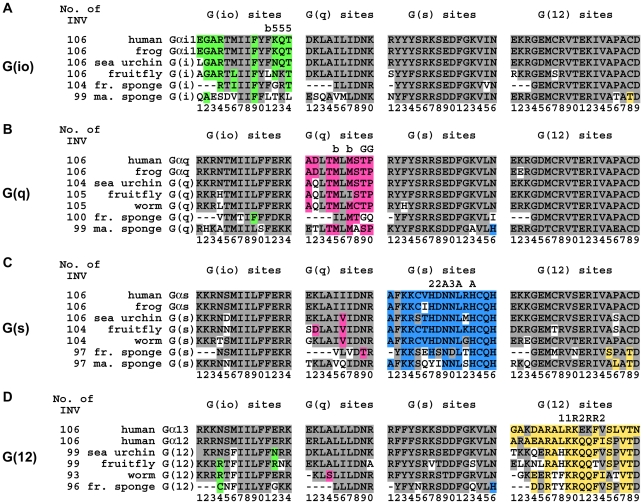
Figure 8. Class-distinctive signature sequences of Gα family members from select organisms.
Signature sequences from select organisms are used to follow the evolution of class-distinctive sites (also see Figure 2). Each panel reflects the evolutionary history of a subtype: (A) Gαi1, (B) Gαq, (C) Gαs, (D) both Gα12 and Gα13. Signature sequences are formed by grouping all distinctive sites for a given class together, removing all residues between individual distinctive sites of the noted class. The distinctive sites for each class are presented in order from the N-terminus to the C-terminus and numbered accordingly. Amino acids that correspond to the ∂ values at that site are colored according to distinctive class: green = G(io); magenta = G(q); blue = G(s); and yellow orange = G(12). Class-distinctive sites were determined using only mammalian sequences. Occasionally a non-mammalian subunit will contain a ∂ or variable (white) amino acid where only η amino acids were observed in the mammalian sequences (for comparison see Figures 4D, 5D, 6D, 7D). (A) Class-distinctive signature sequences of G(io) family members from select organisms. Sites in helix 5 that have been proposed to be important for coupling to the GPCR are denoted by ‘5’ above the site, while site 11 in strand 6 is denoted by ‘b’ above the site. (B) Class-distinctive signature sequences of G(q) family members from select organisms. Sites that interact with GRK2 are denoted by ‘G’ above the site, while sites that are buried and not visible are denoted by ‘b’ above the site. (C) Class-distinctive signature sequences of G(s) family members from select organisms. Sites that have been proposed to be important for the interaction with adenylyl cyclase are denoted by ‘A’ above the site, while additional sites in switches II or III are denoted by ‘2’ or ‘3’, respectively, above the site. (D) Class-distinctive signature sequences of G(12) family members from select organisms. Sites that have direct interactions with p115RhoGEF are denoted by ‘R’ above the site, while additional sites in switches I or II are denoted by ‘1’ or ‘2’, respectively, above the site. Marine sponge = Geodia cydonium; Freshwater sponge = Ephydatia fluviatilis; Worm = Caenorhabditis elegans; Fruitfly = Drosophila melanogaster; Sea urchin = Strongylocentrotus purpuratus; Frog = Xenopus laevis; Human = Homo sapiens.