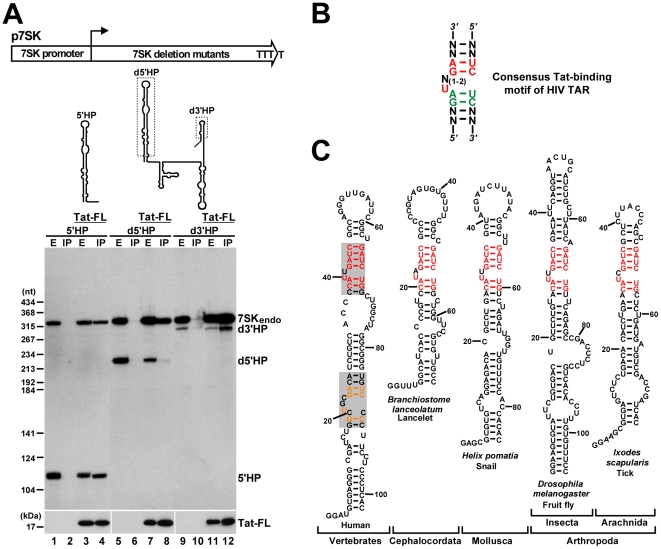
Figure 2. Identification of putative Tat-binding motifs in 7SK snRNA.
A. Tat binds to the 5′ hairpin of 7SK. Schematic structures of the p7SK expression construct and the expressed truncated 7SK RNAs are shown. Dashed boxes indicate deletions. Pol III transcription of the 7SK gene terminates within four consecutive T residues. HeLa cells were transfected with the indicated expression plasmids. After extract (E) preparation, Tat-FL was immunoprecipitated (IP). Distribution of the endogenous and transiently expressed 7SK RNAs and Tat-FL was monitored by Western and Northern blotting. B. Consensus structure of the minimal Tat-binding motif of HIV TAR. Nucleotides with essential and moderate contribution to Tat binding are in red and green, respectively. C. Phylogenetic comparison of the 5′ hairpins of 7SK snRNAs. The sequences of lancelet, snail, fruit fly and tick 7SK snRNAs have been published [65], [66]. The potential Tat-binding motifs of human 7SK snRNA are shaded. The evolutionarily invariant nucleotides are in red. Nucleotides common to the putative proximal Tat-binding motif of 7SK and the Tat-binding site of HIV TAR are in orange.