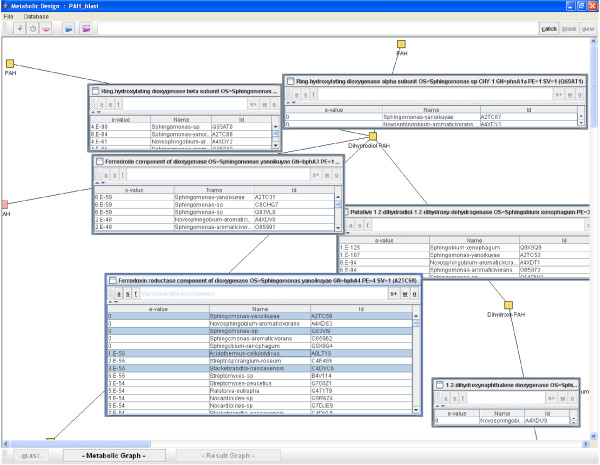
Figure 1.
Results window produced by Metabolic Design. Each metabolite is represented by small yellow squares, called nodes, and enzymes as edges between nodes. Inner windows give the parsing of BLASTp results ordered by increasing Expected Value and obtained for each reference protein as query. For each extracted homologous protein sequence, data such as sequence in EMBL format (f button), sequence in FASTA format (s button), or split BLASTp alignment results (a button) are directly available through the inner window toolbar buttons. The w button, allows the execution of ClustalW alignment on pre-selected protein sequences. Such sequences can also be saved in a single file in FASTA format (s+ button), and/or used to launch the probe design module (o button). Additional functions have also been implemented. The user can automatically highlight potential metabolic capacities of a given organism (species name) subsequently using the 'catch' and 'view' buttons at the top of the window.