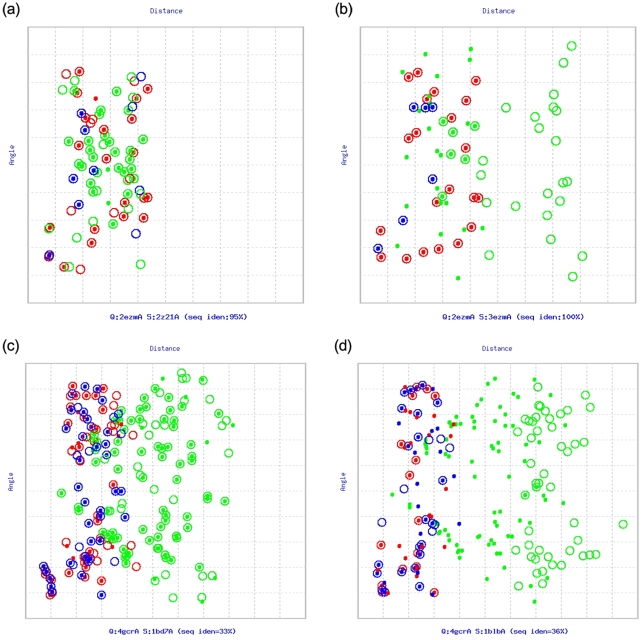
Figure 1. Matched angle-distance (A-D) images.
(a) Two open-form cyanovirin-N molecules (PDB entries: 2ezmA and 2z21A). (b) 3D domain-swapping cyanovirin-N proteins (2ezmA and 3ezmA), a “bona fide” case [2]. (c) γB-crystallin from bovine (4gcrA) and βB2-crystallin from rat (1bd7A), common structural homologs with closed conformations. (d) γB-crystallin from bovine (4gcrA) and an iron-dependent regulator from Mycobacterium tuberculosis (1b1bA), an example of “quasi-domain swapping” [2]. Two secondary structural elements (SSEs), which have been transformed into vectors, in a protein structure form an SSE pair. In these images, the angle of SSE pairs is plotted on the y-axis, and the Euclidean distance of geometric centers of the SSEs is plotted on the x-axis. Both axes have been normalized. The dots and circles represent SSE pairs from the query and subject proteins, respectively. If two SSE pairs from different proteins can be matched (see MATERIALS AND METHODS), they are drawn as a concentric pair of dot and circle. Every protein shown here can be divided into two parts, i.e., a main domain and a swapped domain, within which the SSE pairs are painted red and blue, respectively, while the SSE pairs formed between these (inter-domain SSE pairs) are painted green. Clearly, 3D domain-swapping homologs ((b) and (d)) have a different pattern from common structural homologs ((a) and (c)) in the matched A-D images, where the data points of inter-domain SSE pairs of the open- and closed-form homologs are distributed separately and cannot be well-matched.