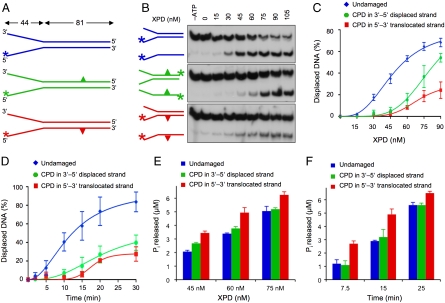
Fig. 3.
Differential impact on XPD enzyme activity. (A) Schematic view of fork substrates. The CPD is located either in the 5′–3′ translocated or the 3′–5′ displaced strand. (B) Typical autoradiographs showing the inhibition of XPD helicase by a single CPD either in the translocated (Bottom) or displaced strand (Middle) of forked substrates (5 nM). (C) Dose dependence of helicase activity. The indicated concentrations of FaXPD were incubated (15 min) with forked DNA substrate (5 nM). The CPD is located either in the translocated or the displaced strand. Oligonucleotide displacement is expressed as the percentage of total radioactivity in each reaction (N = 3; ± SD). (D) Time course experiments. FaXPD (60 nM) was incubated with forked substrates (5 nM) for the indicated time periods (N = 3; ± SD). (E) Dose-dependent stimulation of ATPase activity. The indicated concentrations of FaXPD were incubated with forked DNA (5 nM) in helicase reaction buffer containing 3 mM ATP (N = 3; ± SD). (F) Time course of Pi release upon incubation of FaXPD protein (60 nM) with forked DNA substrates (5 nM) (N = 3; ± SD).