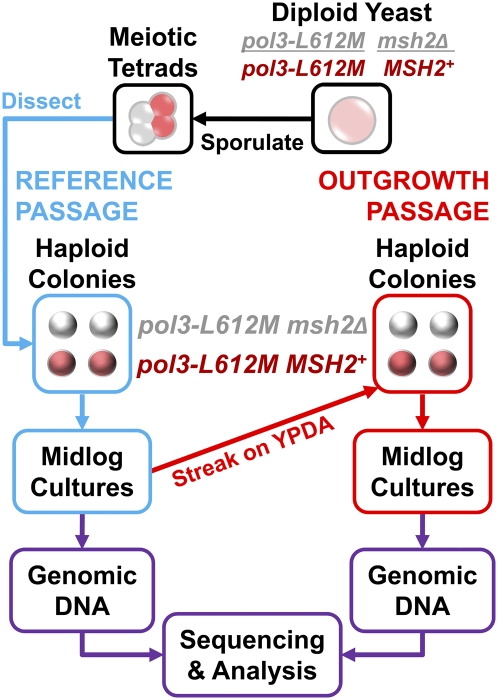
Fig. 2.
Protocol to obtain genomic DNA for sequence analysis. A diploid strain homozygous for pol3-L612M and heterozygous for deletion of MSH2 was sporulated to generate meiotic tetrads. These tetrads were dissected, and colonies resulting from the single-cell meiotic haploid products were grown overnight in 10 mL of YPDA medium. These cultures were added to 90 mL of YPDA medium and grown for 6 h to obtain ≈1010 cells, requiring ≈33 generations. This reference passage (blue path) is the period in which most or all of the mutations to be analyzed were generated. DNA obtained from this first passage, extracted from the whole population and, thus, representing the baseline haploid cells that emerged from tetrad dissection, served as the reference genome for each clone. Single colonies were obtained from these cultures by streaking out on YPDA plates, followed by a second round of growth in liquid YPDA medium. This outgrowth passage (red path) served to isolate and amplify genomes that were subject to mutation during the reference passage. DNA was extracted and sequenced to determine the uncorrected Pol δ L612M replication errors that had accumulated during the first round of growth.