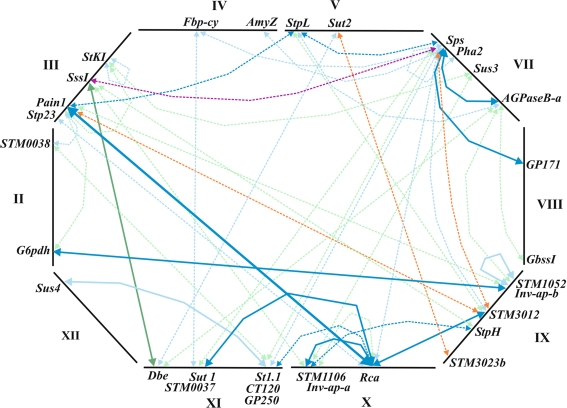
Fig. 2.
Map of pairwise epistatic interactions associated with TSC, TSY and chip quality (CQA or CQS). Marker pairs significant at q ≤ 0.10 and 0.10 < q ≤ 0.20 are connected by solid and broken-line arrows, respectively. Dark blue arrows Associations with TSC and TSY, light blue arrows associations with TSC, blue-green arrows associations with TSY, orange arrows associations with CQA, purple arrow association with TSC, TSY and CQS. Details of the interaction pairs can be found in supplementary Table 1. The positions of the 31 interacting loci on potato chromosomes II–V, VII–XII are shown schematically, for further details see Li et al. (2008). Loci encoding candidate genes are (clock-wise starting with chromosome II): G6pdh cytoplasmic glucose-6-phosphate dehydrogenase, Stp23 plastidic L-type α-glucan phosphorylase, Pain1 vacuolar soluble acid invertase, SssI soluble starch synthase I, StKI clustered gene family of Kunitz-type enzyme inhibitors, Fbp-cy cytoplasmic fructose-1,6-bisphosphatase, AmyZ α-amylase, StpL plastidic l-type starch phosphorylase, Sut2 sucrose sensor or transporter, Sps sucrose phosphate synthase, Pha2 plasma membrane H+ -ATPase 2, Sus3 sucrose synthase 3, AGPaseB-a ADP-glucose pyrophosphorylase, B subunit, GbssI granule-bound starch synthase I, Inv-ap-b tandem duplicated apoplastic invertase genes InvGE and InvGF, StpH cytoplasmic H-type starch phosphorylase, Rca ribulose-bisphosphate carboxylase/oxygenase activase, Inv-ap-a duplicated apoplastic invertase genes, Sut1 sucrose transporter 1, Dbe debranching enzyme, Sus4 sucrose synthase 4. STM0038, STM1052, STM3012, STM3023b, STM1106 and STM0037 are SSR loci. Loci without known function in carbohydrate metabolism are GP171, GP250, CT120 and St1.1