Abstract
Musa (banana and plantain) is an important genus for the global export market and in local markets where it provides staple food for approximately 400 million people. Hybridization and polyploidization of several (sub)species, combined with vegetative propagation and human selection have produced a complex genetic history. We describe the application of the Ecotilling method for the discovery and characterization of nucleotide polymorphisms in diploid and polyploid accessions of Musa. We discovered over 800 novel alleles in 80 accessions. Sequencing and band evaluation shows Ecotilling to be a robust and accurate platform for the discovery of polymorphisms in homologous and homeologous gene targets. In the process of validating the method, we identified two single nucleotide polymorphisms that may be deleterious for the function of a gene putatively important for phototropism. Evaluation of heterozygous polymorphism and haplotype blocks revealed a high level of nucleotide diversity in Musa accessions. We further applied a strategy for the simultaneous discovery of heterozygous and homozygous polymorphisms in diploid accessions to rapidly evaluate nucleotide diversity in accessions of the same genome type. This strategy can be used to develop hypotheses for inheritance patterns of nucleotide polymorphisms within and between genome types. We conclude that Ecotilling is suitable for diversity studies in Musa, that it can be considered for functional genomics studies and as tool in selecting germplasm for traditional and mutation breeding approaches.
Electronic supplementary material
The online version of this article (doi:10.1007/s00122-010-1395-5) contains supplementary material, which is available to authorized users.
Keywords: Banana, Ecotilling, Enzymatic mismatch cleavage, Musa, Nucleotide variation
Introduction
The genus Musa, including edible banana and plantains, ranks in the top 20 food crops in terms of worldwide production and is especially important in developing countries (FAOSTAT 2010). Edible banana and plantain are commonly seedless diploid and triploid hybrids derived from crosses between different subspecies within diploid Musa acuminata containing the A genome and with Musa balbisiana containing the B genome (Simmonds and Shepherd 1955). Because triploid varieties are highly sterile, edible plants are typically propagated asexually. Since banana was domesticated at least 7,000 years before present (Denham et al. 2003), human interventions in the cultivation of banana have occurred for thousands of years (INIBAP 1995). Vegetative maintenance of diploid and triploid clones allowing for novel diversity through the fixation of natural mutation events, along with hybridization of diploids and the creation of new polyploid hybrids, has led to a complex genetic diversity that has yet to be taxonomically fully resolved (Heslop-Harrison and Schwarzacher 2007). It has been estimated that between 1,500 and 3,000 Musa accessions have been collected and a variety of genetic diversity studies have been performed on small non-overlapping subsets of this group of accessions (Heslop-Harrison and Schwarzacher 2007). Many studies have focused on diversity of the nuclear genome using marker techniques including RAPD, AFLP, STMS, IRAP and also methods like genomic in situ hybridization,(GIS; for examples, see (Bhat et al. 1995; D’Hont et al. 2000; Lagoda et al. 1998; Loh et al. 2000; Nair et al. 2005). Such methods provide overall measures of genomic diversity but do not readily provide information on variation at the nucleotide level for gene-coding sequences.
Ecotilling is a high-throughput method for the discovery and characterization of single nucleotide polymorphisms (SNPs) and small insertions/deletions (indels). It is an adaptation of the enzymatic mismatch cleavage and fluorescence detection methods originally developed for the Targeting Induced Local Lesions IN Genomes (TILLING) reverse-genetic strategy (Colbert et al. 2001; Comai et al. 2004). First described for Arabidopsis ecotypes (hence Ecotilling) it has since been shown to be an accurate, low-cost and high-throughput method for the discovery and evaluation of nucleotide diversity in humans, switchgrass, poplar, melon and other organisms (Gilchrist et al. 2006; Nieto et al. 2007; Till et al. 2006a; Weil 2009).
For Ecotilling using enzymatic mismatch cleavage, ~700–1,600 bp gene target regions are amplified using gene-specific primers that are fluorescently labelled. After PCR, samples are denatured and annealed, and heteroduplexed molecules are created through the hybridization of polymorphic amplicons. Mismatched regions in otherwise double-stranded duplex are then cleaved using a crude extract of celery juice containing the single-strand specific nuclease CEL I. Cleaved products are resolved by denaturing polyacrylamide gel electrophoresis (PAGE) and observed by fluorescence detection (Till et al. 2006b). Denaturing PAGE provides base pair resolution allowing grouping of accessions based on shared banding patterns indicative of haplotype grouping (Comai et al. 2004). Sequence validation can be performed on only one or a small number of samples to provide base polymorphism data for the whole group, providing a savings in cost and informatics load over sequencing approaches. Alternatively, banding patterns alone can be used to evaluate genetic diversity and similarity between accessions on a gene-specific scale. When samples are screened alone, Ecotilling provides a catalogue of heterozygous nucleotide diversity between samples. Reference DNA can be added to each sample prior to screening to uncover homozygous polymorphisms. Additionally, the high sensitivity of the assay allows for pooling of multiple samples for the specific discovery of rare polymorphisms as has been described for human samples where minor alleles can contribute to complex polygenic diseases and cancer (Till et al. 2006a).
We describe here the adaptation and application of Ecotilling for polymorphism discovery in Musa. We choose 80 accessions from the International Musa Germplasm Collection, a sample set that serves as a common reference for diversity studies and technology development in banana and plantain. From this work we estimate errors to be low relative to resequencing strategies, and similar to accuracies previously reported for Ecotilling for human polymorphisms. We describe applications for genomic diversity studies and germplasm characterization and further discuss Ecotilling for functional gene analysis and reverse-genetic strategies using TILLING.
Materials and methods
DNA extraction
Plants used for this study were obtained from the Bioversity’s International Transit Centre (ITC, http://www.crop-diversity.org/banana/#AvailableITCAccessions), or from internal collections of IAEA (Supplementary file 1). Genomic DNA samples were prepared from leaf tissue using the DNeasy plant mini kit (QIAGEN). Samples were evaluated for quality and quantity using a standard agarose gel assay as previously described (Till et al. 2006b). Samples were normalized to a concentration of ~0.02 ng/μl prior to PCR amplification.
Primer design and Ecotilling
BAC sequence from the Musa Genome Project was used to design primers to amplify approximately 750–1,500 bp gene-coding target regions (Table 1). Target regions ACETRANS, ADPGP, BETAMHD, DNAJ, FTSJ, GTPFP, HP1, HP2, LPTR, NPH3, RHP, SCPD, STARCHST and WRKY were chosen using the CODDLE software and primers designed using Primer3 with a minimum of 67°C and maximum of 73°C melting temperature (McCallum et al. 2000; Rozen and Skaletsky 2000). Forward primers were labelled with IRDye700 dye and reverse primers with IRDye800 dye. PCR amplification was performed using 0.1 ng of DNA per reaction. PCR amplification, enzymatic mismatch cleavage using a crude celery juice extract, denaturing polyacrylamide gel electrophoresis and fluorescence detection using the LI-COR DNA analyzer were as previously described (Till et al. 2006b). For standard runs, samples were not pooled prior to screening and only heterozygous polymorphisms were discovered. An equal amount of ‘Calcutta4’ reference DNA was added to test samples prior to PCR to uncover homozygous nucleotide differences.
Table 1.
Gene targets, primer sequences and amplicon lengths used in Ecotilling
| Target name | GenBank locus | Locus tag | Forward primer (5′–3′) | Reverse primer (5′–3′) | Amplicon size (bp) |
|---|---|---|---|---|---|
| ACETRANSa | AC186756 | MA4_112I10.20 | TCGCTCTGGGTTTCAGGAAAGCAGTT | TCAGAGTGTAAACCGGGGTCCCAAAT | 1,500 |
| ADPGP | AC186746 | MA4_25J11.14 | GTCAGGCAATTCACTTGGGTA | GGTGCCTCATGTTGATGATTC | 903 |
| BETAMHDa | AC186746 | MA4_25J11.12 | GTTGAGGCTGCAGGAGGAGAGTTCATT | TTCTCTCTCATGGCCTTTGCCTTCTTG | 1,499 |
| DNAJa | AC186747 | MA4_106O17.46 | AGGAGAAGTCAGGGACCAGAACCGAAT | TATAAACCGCCCAAATCTCACCACAGC | 1,473 |
| FTSJ | AC186746 | MA4_25J11.4 | TGGTACCTTTGGATGAGGATG | CTGGGTGATTTTGCAGATTGT | 901 |
| GTPFP | AC186746 | MA4_25J11.8 | GTGGCACTAAATTGGGTCAAA | GGAACTGGCAGAGGAAAAACT | 903 |
| HP1 | AC186746 | MA4_25J11.28 | CCAGCAGTTCATGGACCTTTA | CTGCCTTCCATAGCTCATCTG | 901 |
| HP2 | AC186746 | MA4_25J11.10 | CCTGAGCACTACCAAGGATCA | ATTCCCATGTCATCTCGTCAC | 895 |
| LPTRa | AC186747 | MA4_106O17.14 | GCTCTGACCTCAGTCCCATCACCTCTT | TTCTTGCTCGATCATGGTGACATTCAT | 745 |
| NPH3a | AC186747 | MA4_106O17.30 | TCGAACCTGCTGCCAAGTTCTGTTATG | GTCCATGCTCACCTTCAAGACCTGGTT | 1,498 |
| RHP | AC186748 | MA4_8L21.22 | TGGCCCCTTAACTAAGCATCT | CCTTGGAAGGAAACTGAAAGG | 899 |
| SCPDa | AC186753 | MA4_54B05.24 | GGATTGCAAGTTCTCTTGGCATGCTTCAT | TTGGACATCCGGTCGGTTGAAATACAC | 1,500 |
| STARCHSTa | AC186747 | MA4_106O17.2 | TGAAAATGGTCATAAGCTGTTTGAGACG | CTATTTCCATGATGTGGCTTCGGATGA | 1,496 |
| WRKY | AC186746 | MA4_25J11.46 | GGGAGAGAGTAGCGAGCCTTA | AGAAGGTGTGCGATTCGATTA | 896 |
aTargets used in replicate screening
Data analysis and sequence confirmation
Tiff gel images produced by the LI-COR DNA analyzer were manually scored using the GelBuddy program (Zerr and Henikoff 2005). For each gel run, an analysis window smaller than the target amplicon size was manually chosen based on image quality and the absence of PCR mispriming artefacts that can occur near the primer-binding region (Till et al. 2006b). All bands in the selected region were scored. Data summary reports produced in GelBuddy were imported into Microsoft Excel for further analysis. A two-dimensional map was generated for each gel run that notes the presence (indicated as a 1), absence (0) of a band at a specific molecular weight where at least one band was observed for ≥one accession, or the failure of the lane (?). Signals in replicate sample lanes and failed lanes were counted using the IF/AND Excel formula. Samples sharing the same banding pattern were grouped by binning samples according to the sum of the molecular weights of all polymorphic bands. Percent heterozygosity was calculated as the number of bands identified per nucleotide. For example, if 10 bands are identified in a 1,000 bp analysis window, a heterozygosity of 1% (10/1,000 × 100) is reported. To compare heterozygosity measurements between diploid AA and triploid AAA samples, a two-tailed t test with unequal variance was performed using Microsoft Excel for the data set from gene targets ACETRANS, BETAMHD, DNAJ, LPTR, NPH3, SCPD, and STARCHST. Principal component analysis was performed using MVSP (Multi-Variate Statistical Package version 3.13, Kovach Computing Services, Wales) and default settings. Analysis was performed with data collected using the same primers as the two-tailed t test. Accessions Honduras (BB), Tani (BB), Zebrina (AA) and FHIA-01 (AAAB) were excluded because both replicates contained failed lanes in some of the gene targets. For all other accessions, a replicate with no failed lanes was included in the analysis. Nucleotide polymorphisms were confirmed by Sanger sequencing as previously described (Till et al. 2003). The potential effect of SNPs on protein function was evaluated using the SIFT and PARSESNP programs (Ng and Henikoff 2003; Taylor and Greene 2003).
Results
To evaluate the nucleotide diversity in Musa, we chose 80 accessions representing diploid and polyploids mainly of the A and B genome types. Oligonucleotide primers amplifying 14 gene-containing amplicons of 745 bp to 1.5 kb were designed from BAC sequences (Table 1). Ecotilling assays were performed using enzymatic mismatch cleavage and fluorescence detection as previously described (Fig. 1; Till et al. 2006b). To evaluate the efficiency of Ecotilling polymorphism discovery in Musa, 48 diploid and triploid accessions of the A and B genome type were screened using 7 gene-specific primer pairs (Table 1; Supplementary file 1). A technical replicate (identical DNA preparation) for each sample was included in the 96-lane assays. The presence or absence of bands in replicated samples was used to estimate discovery errors. Analysis revealed 93% (2,882/3,105) of bands replicated in the two technical replicates of each genotype (Supplementary file 2). This count includes gel lanes marked as failures, presumably due to PCR amplification failure or human error, and is therefore an estimate of the total error in the assay. When corrected for failed lanes, we estimate an accuracy of 98% (2,882/3,105–172). This estimate counts bands found in only one of two sample lanes and therefore combines false discovery and false-negative errors. Additional false-negative errors where a band is missing from both replicate samples are unknown. We expect, however, that such errors are rare because a large-scale Ecotilling study with human samples revealed a 4% false discovery and 5% false-negative rate when screening unpooled samples as compared to polymorphism data collected by Sanger sequencing (Till et al. 2006a). To further evaluate accuracy of Musa Ecotilling, we compared discovered bands with sequence data from gene target NPH3 that is homologous to the Arabidopsis gene nonphototropic hypocotyl 3 involved in phototropism (Motchoulski and Liscum (1999); Table 2). Of 16 SNPs discovered by sequencing full amplicons of all ten accessions, one heterozygous nucleotide change was not identified in the Ecotilling assay. While the sample size is small, this false-negative rate (~6%, 1/16) is similar to the 5% reported for Ecotilling in human samples with 171 evaluated SNPs (Till et al. 2006a). No sequence change could be validated for one band discovered by Ecotilling, suggesting a similarly low false discovery error rate. Sequencing revealed only heterozygous SNPs at Ecotilling band positions. This is expected when performing enzymatic mismatch cleavage on unpooled DNA samples. Homozygous changes go undetected in Ecotilling reactions unless a reference sample is added to create heteroduplexed molecules that are the substrate for enzymatic mismatch cleavage. In cases where the same molecular weight Ecotilling band was observed in multiple accessions, all accessions shared the same nucleotide change (4/4). Of 13 unique nucleotide changes confirmed by sequencing, six were silent synonymous changes and the remaining seven resulted in non-silent missense changes. Two missense changes are predicted to be deleterious to protein function using the SIFT or PARSESNP programs (Table 2).
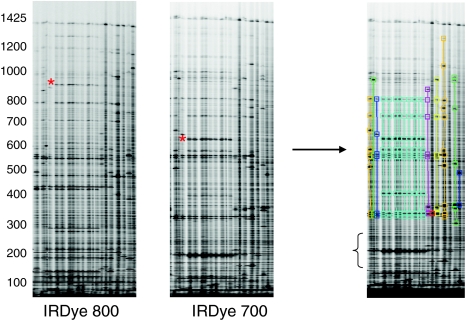
Fig. 1.
Polymorphism discovery in Musa by Ecotilling. IRDye 800 and IRDye 700 images shown for 20 lanes of a 96 lane assay screening for polymorphisms in the 1,495 bp STARCHST gene target. Data is analysed using the GelBuddy program (Zerr and Henikoff 2005). True nucleotide polymorphisms produce cleaved fragments in each fluorescent channel whose molecular weights sum to the approximate molecular weight of the uncut PCR product (an example is marked with a red asterisk). Molecular weights are provided by the GelBuddy program. Samples with similar banding patterns are recorded as having the same haplotype pattern (right panel samples with the same banding pattern are visually grouped by color, numerical data table not shown). Band selection was performed manually. Putative polymorphisms in gel regions with high levels of noise from primer mispriming (denoted by bracket), and the corresponding fragments in the alternative image channels could not be unambiguously assigned and therefore were not scored
Table 2.
Sequence validation of polymorphisms identified in NPH3 target
| Nucleotide changea | Bandb | Effect | Genotype | SIFTc | PARSESNPd |
|---|---|---|---|---|---|
| C1045S | + | A475G | Leite | 0.13 | 9.4 |
| G867 K | + | V416L | FHIA01 | 1.00 | |
| G860R | + | E413= | Leite, Pisang Kayu, Mbwazirume | ||
| T797 W | + | I392= | FHIA_01 | ||
| C701Y | + | T360= | FHIA-01 | ||
| A672 W | + | T351S | Pahang | 0.42 | 10.8 |
| G641Re | + | K340= | FHIA-01 | ||
| A640Re | + | K340R | FHIA-01 | 0.59 | −0.6 |
| T576Y | + | S319P | Calcutta4 | 0.30 | |
| G515 K | + | R298S | FHIA-01 | 0.03 | 11.2 |
| G494R | + | G291= | FHIA-01 | ||
| G418S | + | C266S | FHIA-01 | 0.41 | 6.4 |
| T377 W | + | P252= | FHIA-01 | ||
| NDb | + (245 bp) | NA | Calcutta4 | ||
| Total (14) | 14 | 16 |
aNucleotide position on PCR amplicon
bAll nucleotide changes identified by sequencing were first identified by Ecotilling as a band on the gel image. In one case a 245-bp band was identified on the Ecotilling gel for which a corresponding polymorphism could not be confirmed by sequencing
cA non-synonymous SNP is predicted to be damaging to the encoded protein if the SIFT score is <0.05 (bold)
dA non-synonymous SNP is predicted to be damaging to the encoded protein if the PARSESNP score is >10 (bold)
eAdjacent polymorphisms appear as a single band on the gel image
We next applied Ecotilling band analysis to the remaining accessions using seven additional gene targets. In total, 6,064 polymorphisms were discovered in 80 accessions and 14 gene targets, for a total of 870 unique alleles (Supplementary file 2). Thus, with only a small number of gel runs, a large number of new genetic markers anchored to genes could be discovered. To evaluate nucleotide diversity between accessions, nucleotide heterozygosity was calculated as the percentage of polymorphic sites (number of Ecotilling bands per sample/total bases screened; Supplementary file 3). Heterozygosity in accessions ranged between 0.1 and 2.9% for the gene targets investigated. Comparison of AA and AAA samples using the t test suggests that heterozygosity differences between the two populations are greater than expected by random chance (P < 0.001; Supplementary file 3). To further evaluate genetic similarities between accessions, banding patterns were used to assign samples into groups containing common polymorphism patterns (haplotype groups; Fig. 1; Supplementary file 4). This was done on a target-by-target basis with 356 unique haplotype groupings catalogued, representing a large genetic diversity in the test accessions. Included in the test samples were six triploid mutant accessions of the ‘Grande Naine’ variety (AAA) that were previously irradiated with gamma-rays. These mutants grouped together by haplotype pattern suggesting genetic similarity. Because all bands found in mutant samples were also found in non-mutagenized accessions, we conclude that no induced mutations are present in the gene fragments assayed. Similarities between accessions using heterozygous SNP position information were further evaluated using principal component analysis (PCA; Fig. 2; Supplementary file 4). This allowed an accurate differentiation (43/44, ~98%) between hybrids of mixed A and B genome types (cluster c; Fig. 2) and accessions harbouring only one genome type (clusters a and b). Triploid ‘Grande Naine’, mutagenized ‘Grand Naine’, and related triploid AAA accessions clustered into a single group. PCA was not sufficient to differentiate between AA and BB accessions, presumably because many distinguishing polymorphisms are homozygous in sexually propagated accessions and therefore not detected when screening individual samples by Ecotilling.
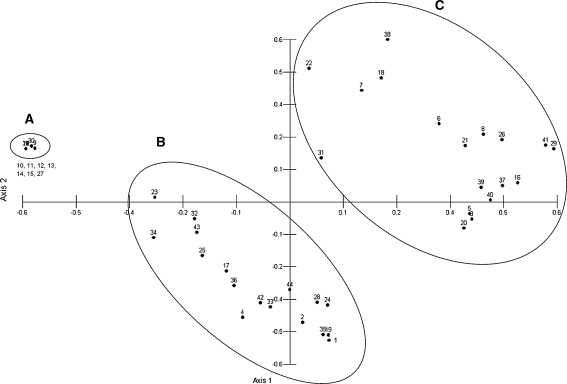
Fig. 2.
Principal component analysis of 44 Musa accessions using SNP position data from 7 gene targets. Three clusters are resolved with ‘Grande Naine’ and related triploid AAA accessions found in cluster a, hybrid accessions of mixed AB chromosomes chromosome type found in cluster c, and accessions of either A or B chromosome type found in cluster b. One sample of mixed chromosome type (Yawa 2, ABBT, sample number 44) was found in cluster b (1/44, 2%). Sample identity listed in Supplementary file 1, PCA table in Supplementary file 4
To uncover homozygous polymorphisms between subject accessions of the M. acuminata genome type and a standard reference, an equal amount of diploid ‘Calcutta4’ (AA) DNA was added to selected test samples prior to PCR amplification. After amplification with primers for the STARCHST gene target, homologous to soluble glycogen starch synthase genes, PCR products were denatured and annealed to form heteroduplexes between ‘Calcutta4’ and polymorphic test sample amplicons. These products were then subjected to enzymatic mismatch cleavage. By screening samples alone and mixed with the reference, one can deduce homozygous polymorphisms existing between samples (Fig. 3; Supplementary file 5). With a large enough sample size, this strategy can provide a simple and high-throughput visual assay for evaluating the potential origins of chromosomal regions. For example, a homozygous polymorphism between the ‘Pisang Mas’ and ‘Calcutta4’ AA varieties was discovered that is detected as a heterozygous polymorphism in the triploid ‘Pisang Kayu’ and ‘Leite’ AAA samples (band 2, Fig. 3). This polymorphism was also detected in ‘Pisang Klutuk Wulung’ (BB) and ‘Pisang Batu’ (BB) accessions suggesting that the nucleotide polymorphism may be of ancient origin and predate the divergence of the two genome types. Similarly, a polymorphism identified in the triploid ‘Pisang Kayu’ (AAA) was not found in any of the A type diploids (band 5, lane H) but identified in the M. balbisiana type diploid ‘Pisang Batu’ (BB) (lane L). To expand on this strategy we exploited the high sensitivity of the Ecotilling method and screened a pool of DNAs containing ten diploid AA varieties. By doing so, in one sample lane we discovered all the nucleotide diversity in a test set of diploids. This revealed a unique polymorphism not found in the three diploid AA samples examined (band 1, lane F). This polymorphism was also observed in ‘Pisang Klutuk Wulung’ (BB) (lane J). Nucleotide polymorphisms were confirmed by Sanger sequencing. Three silent SNPs and one non-synonymous SNP were identified. The non-synonymous change is not predicted to be deleterious to protein function when evaluated using the PARSESNP or SIFT programs (Supplementary file 5).
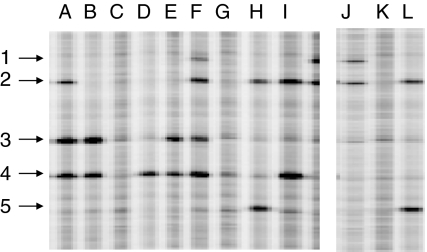
Fig. 3.
Discovery of homozygous nucleotide polymorphisms in diploid Musa acuminata. An equal amount of ‘Calcutta4’ (AA) reference genomic DNA was mixed with ‘Pisang Mas’ (AA) and ‘Pahang’ (AA) prior to PCR with primers for the STARCHST gene fragment to reveal homozygous differences between test and reference samples. Five unique nucleotide polymorphisms were discovered in the gene region shown (numbered 1–5). Samples tested were A ‘Pisang Mas’ + ‘Calcutta4’, B ‘Pahang’ + ‘Calcutta4’, C ‘Pisang Mas’, D ‘Pahang’, E ‘Calcutta4’, F a mixture of 10 diploid AA accessions, G ‘Mbwazirume’ (AAA), H ‘Leite’ (AAA), J ‘Pisang Klutuk Wulung’ (BB), K ‘Honduras’ (BB), L ‘Pisang Batu’ (BB). Polymorphism discovery using a mixture of samples provides a rapid evaluation of diversity within a genome type. Bands were verified as true polymorphisms by Sanger sequencing (Supplementary file 5)
Discussion
Rapid, low-cost and efficient methods for the discovery and characterization of nucleotide polymorphisms in Musa promise to make a positive contribution to understanding genetic diversity and improvement of banana and plantain for food production. Improved crop production is especially important in the context of an increasing population and changing climate. Stresses in food production are likely to increase as populations in developing countries are predicted to rise dramatically, and climate change is expected to adversely affect agricultural production (WDR 2010). Added to this, the edible banana that is a major export and revenue source for developing countries is grown in monoculture and sensitive to a variety of diseases including fungal pathogens causing black sigatoka and panama disease (Marin et al. 2003).
By applying Ecotilling to 14 gene targets and 80 Musa accessions, we have discovered over 6,000 polymorphisms representing 870 unique alleles. Small-scale sequencing validation revealed only SNPs between accessions. While no indels or (micro)satellite repeat variations were sequenced, these are expected at some frequency in the Musa genome. Ecotilling has been previously shown to efficiently recover small indels and repeat variation in plant and human populations, and so we conclude that such nucleotide variation can be readily recovered in Musa (Comai et al. 2004; Till et al. 2006a). A combination of replicate band analysis and sequence confirmation shows that Ecotilling is a highly accurate method for the discovery of nucleotide polymorphisms in Musa, with accuracies similar to those previously achieved for human Ecotilling. That basepair resolution can be achieved, bands of the same molecular weight share the same nucleotide polymorphism, and samples can be grouped according to haplotype pattern allows the consideration of several applications for Musa Ecotilling. The ability to develop high-density SNP patterns anchored to genomic sequences without the need for sequence validation allows for a rapid method to compare and barcode a large number of accessions at relatively low cost. Ecotilling could therefore be used to tag accessions in gene banks, and for the classification of newly acquired samples. Ninety-six accessions can be evaluated in a single gel run and a throughput of 2 to 3 gel runs can be accomplished per day with a single DNA analyzer. Ecotilling of the estimated 3,000 Musa accessions could therefore be accomplished in approximately 11 days per gene target with a single machine. Throughput is scalable with number of machines, and high-throughput TILLING facilities have reported 16 gel runs per day, making multi-gene characterizations of all Musa germplasm feasible (Till et al. 2006a).
Ecotilling was developed as an adaptation of the reverse-genetic technique for the discovery of induced mutations known as TILLING. The same methodologies are used for both TILLING and Ecotilling and therefore the successful adaptation of Ecotilling for Musa suggests that TILLING can also be considered. Indeed, the screening of six gamma-irradiated triploid ‘Grande Naine’ mutants and a non-irradiated ‘Grande Naine’ control can be considered a proof-of-principle for banana TILLING. The mutants were previously selected for increased tolerance to toxin from Mycosphaerella fijiensis (Roux 2001). A total of 9,672 unique bases were screened with the 7 gene target regions used in replicate screening (Table 1). From 6 mutants, a total of 58,032 bases were interrogated for induced mutations. With chemical mutagenesis, mutation density has been shown to vary with ploidy. Data from TILLING projects from a growing number of groups studying diverse plant species are providing baseline data for expectations of observable mutation densities (reviewed in (Till et al. 2009). For example, studies in two-rowed barley suggest variable densities between 1 mutation per 140 and 800 kb depending on ethyl methanesulfonate (EMS) dosage (Gottwald et al. 2009). Higher mutation densities of ~1 mutation per 40 kb were reported for tetraploid wheat (Slade et al. 2005; Till et al. 2003; Uauy et al. 2009). Therefore, one might expect to recover mutations in triploid banana at a density range of 1 mutation per 40 to 140 kb. With only 58 kb screened for mutations in banana, 1 or fewer mutations are expected to be discovered, making the failure to detect any induced mutations unsurprising. Furthermore, mutation density expectations may be inflated because the spectrum (type) of mutations from gamma-irradiation is predicted to be broader than the ≥99% single nucleotide mutations from chemicals such as EMS (Greene et al. 2003; Sato et al. 2006). A greater proportion of deleterious alleles would lower the maximal achievable mutation density, dropping the expected number of discovered mutations in this study to below 1 event. To better evaluate the efficacy of TILLING in banana, a large-scale effort is currently being carried out with an EMS-mutagenized population of ‘Grande Naine’ (Joanna Jankowicz-Cieslak, Chikelu Mba and Bradley J. Till, unpublished). It is especially timely to consider practical reverse-genetic approaches for banana as full genome sequence is expected in the near future (http://www.genoscope.cns.fr/spip/September-8th-2009-Banana-genome.html).
One potentially limiting issue for Ecotilling or TILLING in polyploids is the reported increased false-negative errors when co-amplifying homeologues or related gene targets. Such errors likely arise from competitive hybridization between distinct PCR amplicons followed by a titration of enzymatic mismatch cleavage activity and fluorescence signal due to the presence of many cleavage sites. To avoid this, several methods have been described for homeologue- or copy-specific PCR amplification prior to enzymatic mismatch cleavage (Cooper et al. 2008; Slade et al. 2005). This limitation has been described for TILLING experiments where up to eight unique samples are pooled together prior to PCR, thus lowering the concentration of heteroduplexed molecules arising from induced mutation events. Presumably the discovery of rare mutations present at low concentrations in pools of genomic DNAs from up to eight individual samples is inhibited by natural variations between homeologues or copies. The data presented here suggest that such discovery errors are not present when evaluating high concentration natural polymorphisms between homeologous sequences. We show simultaneous and accurate recovery of polymorphisms from both the A and B genome in diploids and mixed polyploids. This may be due to two main factors: oligonucleotide primers were designed to efficiently amplify homeologues from both A and B genome types, and the concentration of nucleotide polymorphisms is much higher with Ecotilling as compared with highly pooled TILLING samples.
Using Ecotilling for functional gene analysis can also be considered. In this study two potentially deleterious non-silent polymorphisms were identified in the NPH3 gene target that is homologous to genes implicated in the control of hypocotyl length. Indeed, candidate gene approaches have been undertaken to identify natural polymorphisms linked to phenotypes in melon (Nieto et al. 2007). In addition to functional genomics applications, natural allele mining can serve as a primary selection for material to be used in breeding programs. This association genetics or linkage disequilibrium mapping approach is especially appealing in banana due to the lack of mapping populations for QTL studies (Heslop-Harrison and Schwarzacher 2007). It is further strengthened as the sequence of a large number of biologically interesting genes is expected to become available as the genome is being sequenced. For example, parthenocarpy, or seedlessness, is a trait found in edible, diploid, triploid and tetraploid banana. Triploid bananas are susceptible to fungal pathogens such as Mycosphaerella fijiensis, while some seeded diploid subspecies show increased resistance. Ecotilling can be envisioned as a method to identify natural alleles and candidate genes controlling parthenocarpy that can then become targets for traditional breeding, or alternatively as targets for mutation breeding if functional polymorphisms are not available in fertile diploids.
Screening individual DNA samples allowed the rapid and accurate discovery of heterozygous nucleotide polymorphisms. By grouping samples according to common banding patterns, only a small subset of individuals needs to be sequenced to capture the entire nucleotide diversity of a test population. Prior to sequencing, band evaluation can provide important information about the genetic similarity of samples in the test population. Measures of total heterozygosity can be useful in selecting breeding material or a genotype for mutation based approaches such as TILLING. Banding pattern or haplotype analysis provides more information for genetic comparisons and in our study principal component analysis was an accurate way to distinguish samples harbouring both A and B chromosome types from those comprised of only A or B types.
The addition of reference DNA to each sample provides a simple method for the discovery of heterozygous and homozygous polymorphisms. Thus Ecotilling can be used for phylogenetic and population genetic studies that typically employ Sanger sequencing (Gilchrist et al. 2006). The ideal reference sample for Ecotilling is completely isogenic (homozygous) as heterozygous polymorphisms can interfere with the unambiguous assignment of zygosity in test samples. The ‘Calcutta 4’ reference is less than ideal with a measured heterozygosity of ~0.5% (Supplementary file 3). A near homozygous clone has been prepared for genome sequencing through the di-haploidization of the ‘Pahang’ variety (DH-Pahang, ITC accession code: CIRAD930). This is further ideal for Ecotilling as genome sequence and annotation for this clone will become available, making simple the task of mapping sequenced SNPs to test samples and prioritizing the characterization of SNPs in exonic regions for gene-function studies. Enzymatic mismatch cleavage and fluorescence detection was previously shown to be highly sensitive allowing for the efficient discovery of heterozygous mutations in samples pooled up to eightfold (Greene et al. 2003). We utilized this assay sensitivity to develop a strategy for the identification of all nucleotide polymorphisms in a genome type by pooling samples prior to screening. Together, this allows for hypothesis testing of the evolutionary origin of nucleotide polymorphisms. In our study of the STARCHST gene target we identified several polymorphisms found in some, but not all, M. acuminata genotypes that were also identified in M. balbisiana genotypes. This could result from polymorphisms of ancient origin being lost in some A genomes, or through the coincident accumulation of natural mutations. Thus Ecotilling can be used to rapidly identify genomic regions for more extensive analysis. Our study highlights the use of this strategy. The discovery of an acuminata polymorphism pattern in ‘Pisang Klutuk Wulung’ fits with studies suggesting this sample (labelled with ITC code 1063) was mislabeled as ‘Pisang Klutuk Wulung’ (BB) and is in fact an acuminata type (AA) based on morphological studies (Edmond De Langhe, Katholieke Universiteit Leuven, personal communication). These studies also suggested that the ITC1063 sample is likely to be ssp. siamea. This is supported by our PCA analysis where the sample labelled ‘Pisang Klutuk Wulung’ most closely associates with ‘Khae Phrae’, which belongs to ssp. siamea. High sensitivity for low-concentration (rare) polymorphisms in pooled samples has additional advantages. For example, n = 11 (haploid chromosome number) in many diploid accessions but it has been reported that in some triploid ABB genotypes, chromosomal representation deviates from the expected 11A and 22B composition, making sensitivity of SNP discovery by traditional nucleotide sequencing potentially difficult (D’Hont et al. 2000). In cases of skewed chromosomal composition, SNPs can be readily discovered using Ecotilling due to its high assay sensitivity and, if desired, sequence obtained by quantitative massively parallel sequencing.
We conclude that Ecotilling is an accurate and robust method for the discovery and cataloguing of nucleotide polymorphisms in diploid and polyploid accessions of Musa. The technology is highly scalable and many applications can be considered from simple measurements of heterozygosity as a selection criterion in breeding programs to more nuanced studies of chromosomal inheritance and functional genomic analysis. TILLING and Ecotilling core facilities have been developed for plants and animals where a single laboratory services the needs of an entire network of researchers (Till et al. 2009). A similar resource can be envisioned for the Musa community.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
We thank Pierre Lagoda (Plant Breeding and Genetics Section, IAEA) for useful discussions about banana accessions, and Mike Soriano (Chemical Analysis Unit, IAEA) for helpful discussions on the analysis of heterozygosity data. Joy Nakidandwe (IAEA) helped in the design of primers, Danilo Moreno (IAEA), Els Thiry (K.U.Leuven) and Ellen Meelkop (K.U.Leuven) with DNA extraction and first-pass PAGE gel evaluation, and Mirta Matijevic (IAEA) with DNA sequencing. Funding for this work was provided by the Food and Agriculture Organization of the United Nations and the International Atomic Energy Agency through their Joint FAO/IAEA Programme of Nuclear Techniques in Food and Agriculture. We thank the International Musa Germplasm Collection of Bioversity International based at K.U.Leuven for supplying most of the accessions.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Bhat KV, Bhat SR, Chandel PS, Lakhanpaul S, Ali S. DNA fingerprinting of Musa cultivars with oligodeoxyribonucleotide probes specific for simple repeat motifs. Genet Anal. 1995;12:45–51. doi: 10.1016/1050-3862(95)00100-x. [DOI] [PubMed] [Google Scholar]
- Colbert T, Till BJ, Tompa R, Reynolds S, Steine MN, et al. High-throughput screening for induced point mutations. Plant Physiol. 2001;126:480–484. doi: 10.1104/pp.126.2.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comai L, Young K, Till BJ, Reynolds SH, Greene EA, et al. Efficient discovery of DNA polymorphisms in natural populations by Ecotilling. Plant J. 2004;37:778–786. doi: 10.1111/j.0960-7412.2003.01999.x. [DOI] [PubMed] [Google Scholar]
- Cooper JL, Till BJ, Laport RG, Darlow MC, Kleffner JM, et al. TILLING to detect induced mutations in soybean. BMC Plant Biol. 2008;8:9. doi: 10.1186/1471-2229-8-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Hont A, Paget-Goy A, Escoute J, Carreel F. The interspecific genome structure of cultivated banana, Musa spp. revealed by genomic DNA in situ hybridization. Theor Appl Genet. 2000;100:177–183. doi: 10.1007/s001220050024. [DOI] [Google Scholar]
- Denham TP, Haberle SG, Lentfer C, Fullagar R, Field J, et al. Origins of agriculture at Kuk Swamp in the highlands of New Guinea. Science. 2003;301:189–193. doi: 10.1126/science.1085255. [DOI] [PubMed] [Google Scholar]
- FAOSTAT (2010) http://faostat.fao.org/site/339/default.aspx. Accessed 07 January 2010
- Gilchrist EJ, Haughn GW, Ying CC, Otto SP, Zhuang J, et al. Use of Ecotilling as an efficient SNP discovery tool to survey genetic variation in wild populations of Populus trichocarpa. Mol Ecol. 2006;15:1367–1378. doi: 10.1111/j.1365-294X.2006.02885.x. [DOI] [PubMed] [Google Scholar]
- Gottwald S, Bauer P, Komatsuda T, Lundqvist U, Stein N. TILLING in the two-rowed barley cultivar ‘Barke’ reveals preferred sites of functional diversity in the gene HvHox1. BMC Res Notes. 2009;2:258. doi: 10.1186/1756-0500-2-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene EA, Codomo CA, Taylor NE, Henikoff JG, Till BJ, et al. Spectrum of chemically induced mutations from a large-scale reverse-genetic screen in Arabidopsis. Genetics. 2003;164:731–740. doi: 10.1093/genetics/164.2.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heslop-Harrison JS, Schwarzacher T. Domestication, genomics and the future for banana. Ann Bot (Lond) 2007;100:1073–1084. doi: 10.1093/aob/mcm191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- INIBAP (1995) Banana and plantain: the earliest fruit crops? In: INIBAP (ed) INIBAP annual report 1995. INIBAP, Montpellier, pp 6–8
- Lagoda PJ, Noyer JL, Dambier D, Baurens FC, Grapin A, et al. Sequence tagged microsatellite site (STMS) markers in the Musaceae. Mol Ecol. 1998;7:659–663. [PubMed] [Google Scholar]
- Loh JP, Kiew R, Set O, Gan LH, Gan YY. Amplified fragment length polymorphism fingerprinting of 16 banana cultivars (Musa cvs.) Mol Phylogenet Evol. 2000;17:360–366. doi: 10.1006/mpev.2000.0848. [DOI] [PubMed] [Google Scholar]
- Marin DH, Romero RA, Guzman M, Sutton TB. Black sigatoka: an increasing threat to banana cultivation. Plant Dis. 2003;87:208–222. doi: 10.1094/PDIS.2003.87.3.208. [DOI] [PubMed] [Google Scholar]
- McCallum CM, Comai L, Greene EA, Henikoff S. Choosing optimal regions for TILLING. Plant Physiol. 2000;123:439–442. doi: 10.1104/pp.123.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motchoulski A, Liscum E. Arabidopsis NPH3: a NPH1 photoreceptor-interacting protein essential for phototropism. Science. 1999;286:961–964. doi: 10.1126/science.286.5441.961. [DOI] [PubMed] [Google Scholar]
- Nair AS, Teo CH, Schwarzacher T, Harrison PH. Genome classification of banana cultivars from South India using IRAP markers. Euphytica. 2005;144:285–290. doi: 10.1007/s10681-005-7321-2. [DOI] [Google Scholar]
- Ng PC, Henikoff S. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto C, Piron F, Dalmais M, Marco CF, Moriones E, et al. EcoTILLING for the identification of allelic variants of melon eIF4E, a factor that controls virus susceptibility. BMC Plant Biol. 2007;7:34. doi: 10.1186/1471-2229-7-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux NS. Mutation induction in Musa—review. In: Jain SM, Swennen R, editors. Banana improvement: cellular, molecular biology, induced mutations. Enfield: Science Publishers; 2001. pp. 23–32. [Google Scholar]
- Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- Sato Y, Shirasawa K, Takahashi Y, Nishimura M, Nishio T. Mutant selection from progeny of gamma-ray-irradiated rice by DNA heteroduplex cleavage using Brassica petiole extract. Breed Sci. 2006;56:179–183. doi: 10.1270/jsbbs.56.179. [DOI] [Google Scholar]
- Simmonds NW, Shepherd K. Taxonomy and origins of cultivated bananas. J Linn Soc Bot (Lond) 1955;55:302–312. doi: 10.1111/j.1095-8339.1955.tb00015.x. [DOI] [Google Scholar]
- Slade AJ, Fuerstenberg SI, Loeffler D, Steine MN, Facciotti D. A reverse genetic, nontransgenic approach to wheat crop improvement by TILLING. Nat Biotechnol. 2005;23:75–81. doi: 10.1038/nbt1043. [DOI] [PubMed] [Google Scholar]
- Taylor NE, Greene EA. PARSESNP: a tool for the analysis of nucleotide polymorphisms. Nucleic Acids Res. 2003;31:3808–3811. doi: 10.1093/nar/gkg574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Till BJ, Reynolds SH, Greene EA, Codomo CA, Enns LC, et al. Large-scale discovery of induced point mutations with high-throughput TILLING. Genome Res. 2003;13:524–530. doi: 10.1101/gr.977903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Till BJ, Zerr T, Bowers E, Greene EA, Comai L, et al. High-throughput discovery of rare human nucleotide polymorphisms by Ecotilling. Nucleic Acids Res. 2006;34:e99. doi: 10.1093/nar/gkl479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Till BJ, Zerr T, Comai L, Henikoff S. A protocol for TILLING and Ecotilling in plants and animals. Nat Protoc. 2006;1:2465–2477. doi: 10.1038/nprot.2006.329. [DOI] [PubMed] [Google Scholar]
- Till BJ, Afza R, Bado S, Huynh OA, Jankowicz-Cieslak J, et al. Global TILLING projects. In: Shu QY, et al., editors. Induced plant mutations in the genomics era. Rome: Food and Agriculture Organization of the United Nations; 2009. pp. 237–239. [Google Scholar]
- Uauy C, Paraiso F, Colasuonno P, Tran RK, Tsai H, et al. A modified TILLING approach to detect induced mutations in tetraploid and hexaploid wheat. BMC Plant Biol. 2009;9:115. doi: 10.1186/1471-2229-9-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WDR (2010) World Development Report 2010: Development and Climate Change, The World Bank
- Weil CF. TILLING in grass species. Plant Physiol. 2009;149:158–164. doi: 10.1104/pp.108.128785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerr T, Henikoff S. Automated band mapping in electrophoretic gel images using background information. Nucleic Acids Res. 2005;33:2806–2812. doi: 10.1093/nar/gki580. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.