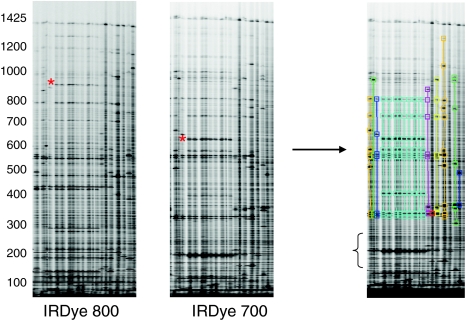
Fig. 1.
Polymorphism discovery in Musa by Ecotilling. IRDye 800 and IRDye 700 images shown for 20 lanes of a 96 lane assay screening for polymorphisms in the 1,495 bp STARCHST gene target. Data is analysed using the GelBuddy program (Zerr and Henikoff 2005). True nucleotide polymorphisms produce cleaved fragments in each fluorescent channel whose molecular weights sum to the approximate molecular weight of the uncut PCR product (an example is marked with a red asterisk). Molecular weights are provided by the GelBuddy program. Samples with similar banding patterns are recorded as having the same haplotype pattern (right panel samples with the same banding pattern are visually grouped by color, numerical data table not shown). Band selection was performed manually. Putative polymorphisms in gel regions with high levels of noise from primer mispriming (denoted by bracket), and the corresponding fragments in the alternative image channels could not be unambiguously assigned and therefore were not scored