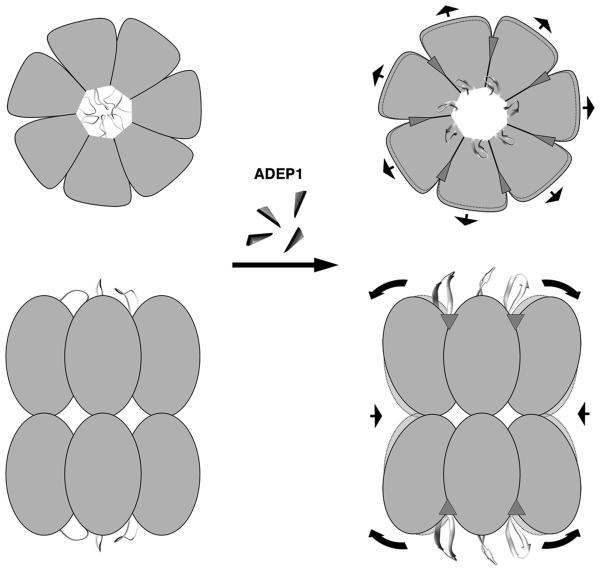
Figure 6. Model for the Activation of ClpP by ADEPs.
Schematic representations of a top and a side-view of ClpP in the absence and presence of ADEP1 are shown in the left and right hand panels, respectively. The N-terminal regions of the ClpP monomers in the absence of ADEP1 are shown in multiple conformations representing the flexible nature of this region and the 12 Å diameter pore that they delimitate. This small diameter pore restricts the passage of protein substrates to the digestion chamber. ADEP1 molecules, represented as small triangles, dock into the seven hydrophobic clefts located on the apical surface of each ClpP ring. Upon binding, ADEP1 locks the ClpP N-terminal loops in a β-hairpin conformation retracting these loops from the lumen and generating a stable pore of 20 Å diameter through which extended polypeptides can be threaded into the degradation chamber. ADEP1 binding also triggers an outward movement of the ClpP head domain causing a subtle expansion of the apical surface of the ring. Simultaneously, the equator of the tetradecamer formed by the ClpP handle domains slightly contracts as a result of the rigid body movement of the ClpP monomers. The arrows indicate the direction of these movements and the areas delimitated by dotted lines represent the ClpP structure before ADEP1 binding and are shown for reference.