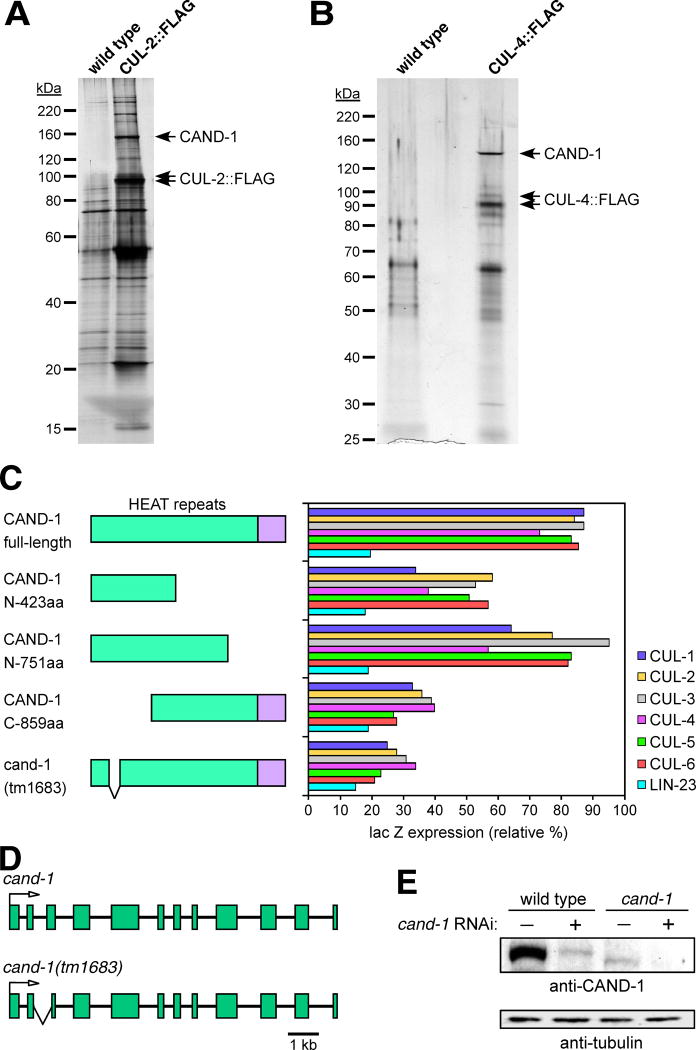
Figure 1. Interaction of CAND-1 with C. elegans cullins.
CAND-1 co-immunoprecipitates with CUL-2∷FLAG (A) and CUL-4∷FLAG (B). Silver-stained SDS-PAGE gels are shown for anti-FLAG affinity purifications from strains containing CUL-2∷FLAG or CUL-4∷FLAG, and from control wild-type animals. The CAND1 protein band (labeled) was identified by mass spectrometry. (C) Two-hybrid analysis of interaction between CAND-1 and the C. elegans cullins. On the left are diagrams of full-length, truncated, and tm1683 mutant CAND-1 proteins (the names reflect the number of N- or C-terminal amino acids remaining in the truncations; the HEAT-repeat region is in green). On the right is a graph of quantitation of interactions between the six cullin proteins and the CAND-1 proteins using a two-hybrid lacZ expression assay. CAND-1 was expressed from the pACT2 vector (fused to the Gal4 activation domain), and cullins or the negative control LIN-23 (Kipreos et al., 2000) are in the pAS1-CYH2 vector (fused to the Gal4 DNA binding domain). The scale derives from the level of the positive control (interaction between pACT2/SKR-1 and pAS1-CYH2/CUL-1; not shown), which is set at 100%. (D) Schematic of the cand-1 genomic region on chromosome V for wild type and the tm1683 deletion mutant. Exons are represented as boxes and lines represent introns. An arrow indicates the translational start point. The region deleted in the tm1683 mutant allele is encompassed by a ‘V-shaped’ lower line. (E) Effect of RNAi on CAND-1 protein levels in wild type and cand-1 mutants. cand-1 RNAi depletion is denoted by a plus above the lanes. Note that cand-1 mutants have lower CAND-1 levels than wild type, and that cand-1 RNAi further reduces CAND-1 levels.