Abstract
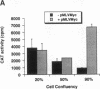
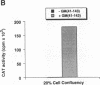
Deregulated expression of the c-Myc oncoprotein occurs in several human malignancies. The c-Myc protein behaves as a transcription factor, and undoubtedly its role in carcinogenesis involves its ability to affect the expression of genes involved in cell growth. c-Myc has been reported to both activate and repress transcription in transient transfection experiments using reporter constructs bearing multiple copies of the c-Myc binding site, CAC (G/A) TG. We investigated these apparently paradoxical effects of c-Myc by determining if they arose from differences in the cell proliferation states of transfected cells. We found that endogenous c-Myc protein levels vary inversely with the degree of cell confluency, such that at low cell confluency, where endogenous levels of c-Myc are high and presumably endogenous levels of Max are limiting, exogenous c-Myc fails to affect basal transcription. In cells at high cell confluency, in which endogenous c-Myc levels are low, exogenous c-Myc augments transactivation by titrating the relative excess endogenous Max. These observations suggest that the apparently paradoxical behavior of c-Myc in transfection experiments is partially dependent on ambient cellular levels of c-Myc.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abken H., Reifenrath B. A procedure to standardize CAT reporter gene assay. Nucleic Acids Res. 1992 Jul 11;20(13):3527–3527. doi: 10.1093/nar/20.13.3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amati B., Brooks M. W., Levy N., Littlewood T. D., Evan G. I., Land H. Oncogenic activity of the c-Myc protein requires dimerization with Max. Cell. 1993 Jan 29;72(2):233–245. doi: 10.1016/0092-8674(93)90663-b. [DOI] [PubMed] [Google Scholar]
- Askew D. S., Ashmun R. A., Simmons B. C., Cleveland J. L. Constitutive c-myc expression in an IL-3-dependent myeloid cell line suppresses cell cycle arrest and accelerates apoptosis. Oncogene. 1991 Oct;6(10):1915–1922. [PubMed] [Google Scholar]
- Auvinen M., Paasinen A., Andersson L. C., Hölttä E. Ornithine decarboxylase activity is critical for cell transformation. Nature. 1992 Nov 26;360(6402):355–358. doi: 10.1038/360355a0. [DOI] [PubMed] [Google Scholar]
- Ayer D. E., Eisenman R. N. A switch from Myc:Max to Mad:Max heterocomplexes accompanies monocyte/macrophage differentiation. Genes Dev. 1993 Nov;7(11):2110–2119. doi: 10.1101/gad.7.11.2110. [DOI] [PubMed] [Google Scholar]
- Ayer D. E., Kretzner L., Eisenman R. N. Mad: a heterodimeric partner for Max that antagonizes Myc transcriptional activity. Cell. 1993 Jan 29;72(2):211–222. doi: 10.1016/0092-8674(93)90661-9. [DOI] [PubMed] [Google Scholar]
- Barrett J., Birrer M. J., Kato G. J., Dosaka-Akita H., Dang C. V. Activation domains of L-Myc and c-Myc determine their transforming potencies in rat embryo cells. Mol Cell Biol. 1992 Jul;12(7):3130–3137. doi: 10.1128/mcb.12.7.3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bello-Fernandez C., Packham G., Cleveland J. L. The ornithine decarboxylase gene is a transcriptional target of c-Myc. Proc Natl Acad Sci U S A. 1993 Aug 15;90(16):7804–7808. doi: 10.1073/pnas.90.16.7804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell T. K., Kretzner L., Blackwood E. M., Eisenman R. N., Weintraub H. Sequence-specific DNA binding by the c-Myc protein. Science. 1990 Nov 23;250(4984):1149–1151. doi: 10.1126/science.2251503. [DOI] [PubMed] [Google Scholar]
- Blackwood E. M., Eisenman R. N. Max: a helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science. 1991 Mar 8;251(4998):1211–1217. doi: 10.1126/science.2006410. [DOI] [PubMed] [Google Scholar]
- Blackwood E. M., Lüscher B., Eisenman R. N. Myc and Max associate in vivo. Genes Dev. 1992 Jan;6(1):71–80. doi: 10.1101/gad.6.1.71. [DOI] [PubMed] [Google Scholar]
- Dang C. V., Barrett J., Villa-Garcia M., Resar L. M., Kato G. J., Fearon E. R. Intracellular leucine zipper interactions suggest c-Myc hetero-oligomerization. Mol Cell Biol. 1991 Feb;11(2):954–962. doi: 10.1128/mcb.11.2.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilers M., Schirm S., Bishop J. M. The MYC protein activates transcription of the alpha-prothymosin gene. EMBO J. 1991 Jan;10(1):133–141. doi: 10.1002/j.1460-2075.1991.tb07929.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evan G. I., Wyllie A. H., Gilbert C. S., Littlewood T. D., Land H., Brooks M., Waters C. M., Penn L. Z., Hancock D. C. Induction of apoptosis in fibroblasts by c-myc protein. Cell. 1992 Apr 3;69(1):119–128. doi: 10.1016/0092-8674(92)90123-t. [DOI] [PubMed] [Google Scholar]
- Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967 Jun 14;26(2):365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- Jansen-Dürr P., Meichle A., Steiner P., Pagano M., Finke K., Botz J., Wessbecher J., Draetta G., Eilers M. Differential modulation of cyclin gene expression by MYC. Proc Natl Acad Sci U S A. 1993 Apr 15;90(8):3685–3689. doi: 10.1073/pnas.90.8.3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaddurah-Daouk R., Greene J. M., Baldwin A. S., Jr, Kingston R. E. Activation and repression of mammalian gene expression by the c-myc protein. Genes Dev. 1987 Jun;1(4):347–357. doi: 10.1101/gad.1.4.347. [DOI] [PubMed] [Google Scholar]
- Karn J., Watson J. V., Lowe A. D., Green S. M., Vedeckis W. Regulation of cell cycle duration by c-myc levels. Oncogene. 1989 Jun;4(6):773–787. [PubMed] [Google Scholar]
- Kato G. J., Barrett J., Villa-Garcia M., Dang C. V. An amino-terminal c-myc domain required for neoplastic transformation activates transcription. Mol Cell Biol. 1990 Nov;10(11):5914–5920. doi: 10.1128/mcb.10.11.5914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato G. J., Dang C. V. Function of the c-Myc oncoprotein. FASEB J. 1992 Sep;6(12):3065–3072. doi: 10.1096/fasebj.6.12.1521738. [DOI] [PubMed] [Google Scholar]
- Kretzner L., Blackwood E. M., Eisenman R. N. Myc and Max proteins possess distinct transcriptional activities. Nature. 1992 Oct 1;359(6394):426–429. doi: 10.1038/359426a0. [DOI] [PubMed] [Google Scholar]
- Land H., Parada L. F., Weinberg R. A. Tumorigenic conversion of primary embryo fibroblasts requires at least two cooperating oncogenes. Nature. 1983 Aug 18;304(5927):596–602. doi: 10.1038/304596a0. [DOI] [PubMed] [Google Scholar]
- Larsson L. G., Pettersson M., Oberg F., Nilsson K., Lüscher B. Expression of mad, mxi1, max and c-myc during induced differentiation of hematopoietic cells: opposite regulation of mad and c-myc. Oncogene. 1994 Apr;9(4):1247–1252. [PubMed] [Google Scholar]
- Lee W. M., Schwab M., Westaway D., Varmus H. E. Augmented expression of normal c-myc is sufficient for cotransformation of rat embryo cells with a mutant ras gene. Mol Cell Biol. 1985 Dec;5(12):3345–3356. doi: 10.1128/mcb.5.12.3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L. H., Nerlov C., Prendergast G., MacGregor D., Ziff E. B. c-Myc represses transcription in vivo by a novel mechanism dependent on the initiator element and Myc box II. EMBO J. 1994 Sep 1;13(17):4070–4079. doi: 10.1002/j.1460-2075.1994.tb06724.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philipp A., Schneider A., Väsrik I., Finke K., Xiong Y., Beach D., Alitalo K., Eilers M. Repression of cyclin D1: a novel function of MYC. Mol Cell Biol. 1994 Jun;14(6):4032–4043. doi: 10.1128/mcb.14.6.4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prendergast G. C., Lawe D., Ziff E. B. Association of Myn, the murine homolog of max, with c-Myc stimulates methylation-sensitive DNA binding and ras cotransformation. Cell. 1991 May 3;65(3):395–407. doi: 10.1016/0092-8674(91)90457-a. [DOI] [PubMed] [Google Scholar]
- Reddy C. D., Dasgupta P., Saikumar P., Dudek H., Rauscher F. J., 3rd, Reddy E. P. Mutational analysis of Max: role of basic, helix-loop-helix/leucine zipper domains in DNA binding, dimerization and regulation of Myc-mediated transcriptional activation. Oncogene. 1992 Oct;7(10):2085–2092. [PubMed] [Google Scholar]
- Roy A. L., Carruthers C., Gutjahr T., Roeder R. G. Direct role for Myc in transcription initiation mediated by interactions with TFII-I. Nature. 1993 Sep 23;365(6444):359–361. doi: 10.1038/365359a0. [DOI] [PubMed] [Google Scholar]
- Sarid J., Halazonetis T. D., Murphy W., Leder P. Evolutionarily conserved regions of the human c-myc protein can be uncoupled from transforming activity. Proc Natl Acad Sci U S A. 1987 Jan;84(1):170–173. doi: 10.1073/pnas.84.1.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seed B., Sheen J. Y. A simple phase-extraction assay for chloramphenicol acyltransferase activity. Gene. 1988 Jul 30;67(2):271–277. doi: 10.1016/0378-1119(88)90403-9. [DOI] [PubMed] [Google Scholar]
- Shi Y., Glynn J. M., Guilbert L. J., Cotter T. G., Bissonnette R. P., Green D. R. Role for c-myc in activation-induced apoptotic cell death in T cell hybridomas. Science. 1992 Jul 10;257(5067):212–214. doi: 10.1126/science.1378649. [DOI] [PubMed] [Google Scholar]
- Shibuya H., Yoneyama M., Ninomiya-Tsuji J., Matsumoto K., Taniguchi T. IL-2 and EGF receptors stimulate the hematopoietic cell cycle via different signaling pathways: demonstration of a novel role for c-myc. Cell. 1992 Jul 10;70(1):57–67. doi: 10.1016/0092-8674(92)90533-i. [DOI] [PubMed] [Google Scholar]
- Stone J., de Lange T., Ramsay G., Jakobovits E., Bishop J. M., Varmus H., Lee W. Definition of regions in human c-myc that are involved in transformation and nuclear localization. Mol Cell Biol. 1987 May;7(5):1697–1709. doi: 10.1128/mcb.7.5.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Antwerp M. E., Chen D. G., Chang C., Prochownik E. V. A point mutation in the MyoD basic domain imparts c-Myc-like properties. Proc Natl Acad Sci U S A. 1992 Oct 1;89(19):9010–9014. doi: 10.1073/pnas.89.19.9010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner A. J., Le Beau M. M., Diaz M. O., Hay N. Expression, regulation, and chromosomal localization of the Max gene. Proc Natl Acad Sci U S A. 1992 Apr 1;89(7):3111–3115. doi: 10.1073/pnas.89.7.3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zervos A. S., Gyuris J., Brent R. Mxi1, a protein that specifically interacts with Max to bind Myc-Max recognition sites. Cell. 1993 Jan 29;72(2):223–232. doi: 10.1016/0092-8674(93)90662-a. [DOI] [PubMed] [Google Scholar]
- Zimmerman K. A., Yancopoulos G. D., Collum R. G., Smith R. K., Kohl N. E., Denis K. A., Nau M. M., Witte O. N., Toran-Allerand D., Gee C. E. Differential expression of myc family genes during murine development. 1986 Feb 27-Mar 5Nature. 319(6056):780–783. doi: 10.1038/319780a0. [DOI] [PubMed] [Google Scholar]