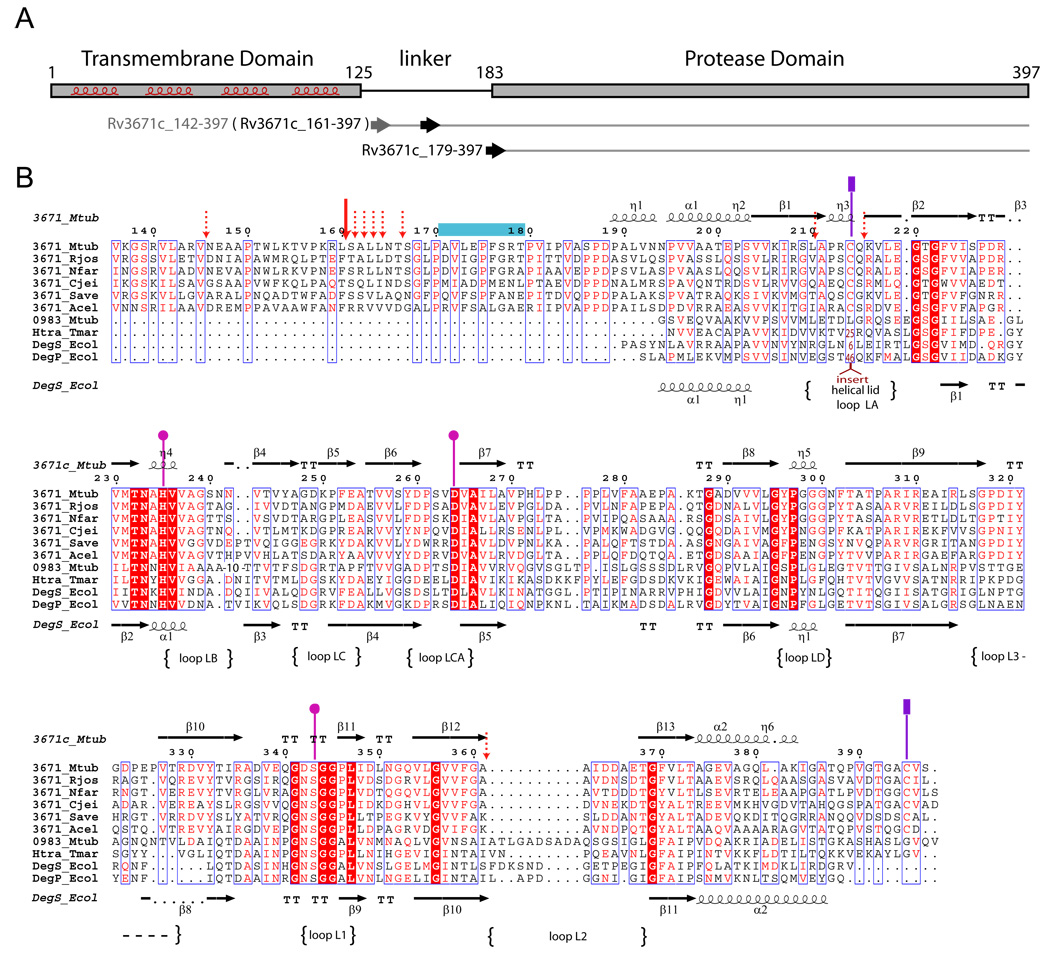
Figure 1.
A. Domain organization of the Rv3671c protein. The predicted transmembrane helices are highlighted. The horizontal arrows indicate the first residues of the recombinant protein constructs (residues 142 and 179) and the first residue (residue 161) of the autocleavage product of Rv3671c_142-397. B. Multiple sequence alignment of Rv3671c homologs from Rhodococcus jostii RHA1 (Rjos), Nocardia farcinica IFM 10152 (Nfar), Corynebacterium jeikeium K411 (Cjei), Streptomyces avermitilis MA-4680 (Save), Acidothermus cellulolyticus 11B (Acel) and four HtrA homologs: DegS and DegP from Escherichia coli K12 (Ecol), Rv0983 from Mycobacterium tuberculosis H37Rv (Mtub), HtrA from T. maritima (Tmar). The residues of the catalytic triad are designated by pink circles. The cysteine residues of the disulfide bond are marked by purple rectangles. The dominant and minor autocleavage sites (as determined by MALDI-MS analysis) are marked by the solid and dashed red arrows, respectively. The peptidic substrate mimic bound to the active site in the crystal is denoted by a blue horizontal bar. The loop names correspond to the serine protease nomenclature (Perona and Craik, 1995).
See also Figure S1.