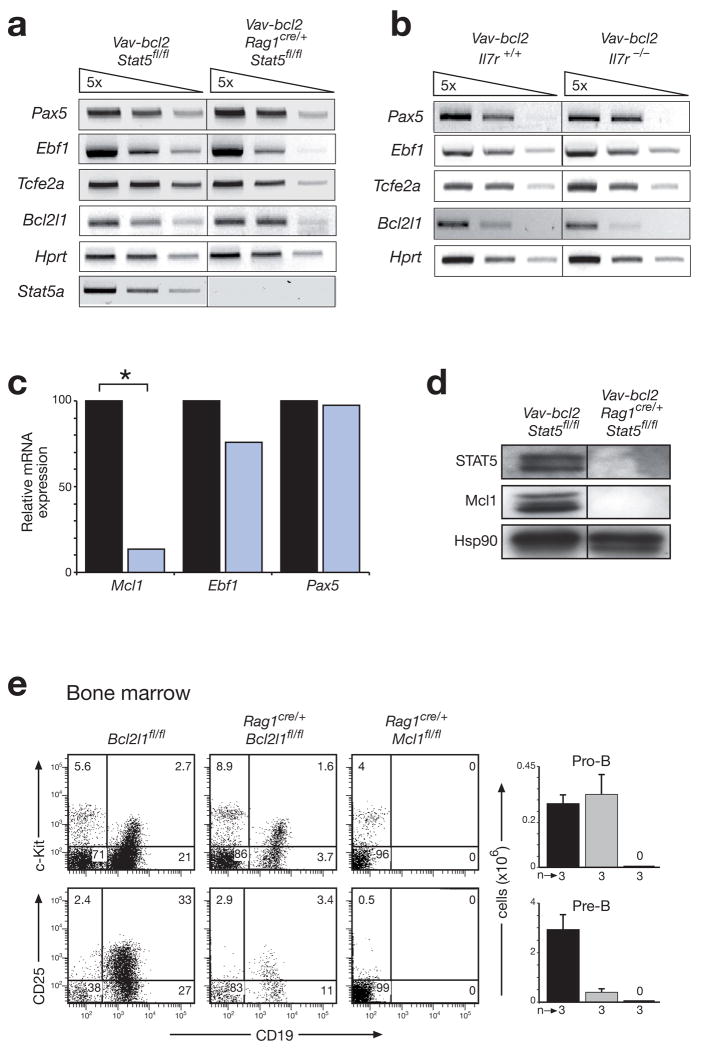
Figure 3. STAT5 controls the survival of pro-B cells by activating the Mcl1 gene.
(a,b) Normal expression of B cell transcription factor genes in the absence of STAT5 (a) or IL-7R signaling (b). Expression of the indicated genes was determined by semiquantitative RT-PCR analysis using 5-fold serial dilutions of cDNA prepared from FACS-sorted pro-B cells (Supplementary Fig. 10) of 4-6-week-old mice of the indicated genotypes. Tcfe2a (E2A); Bcl2l1 (Bcl-x). (c) Reduced Mcl1 expression in STAT5-deficient pro-B cells. Transcripts of the indicated genes were analyzed by real-time RT-PCR of cDNA prepared from sorted pro-B cells of control Vav-bcl2 Stat5fl/fl (black bar) and Vav-bcl2 Rag1cre/+ Stat5fl/fl (blue bar) mice. The transcript levels of control pro-B cells were set to 100%. Statistical significance was determined by the student’s t test (*, p < 0.05). (d) Loss of Mcl-1 expression in the absence of STAT5. Whole cell extracts of sorted CD19+ bone marrow cells from mice of the indicated genotypes were analyzed by immunoblot analysis with STAT5, Mcl-1 and Hsp90 antibodies. (e) Distinct functions of Mcl1 and Bcl2l1 in early B cell development. Pro-B and pre-B cells were analyzed by flow cytometric analysis in the bone marrow of control Bcl2l1fl/fl (black bar), Rag1cre/+ Bcl2l1fl/fl (grey bar) and Rag1cre/+ Mcl1fl/fl mice (denoted by 0). Absolute cell numbers are shown to the right together with the number (n) of mice analyzed. A more comprehensive FACS analysis is shown in Supplementary Fig. 12.