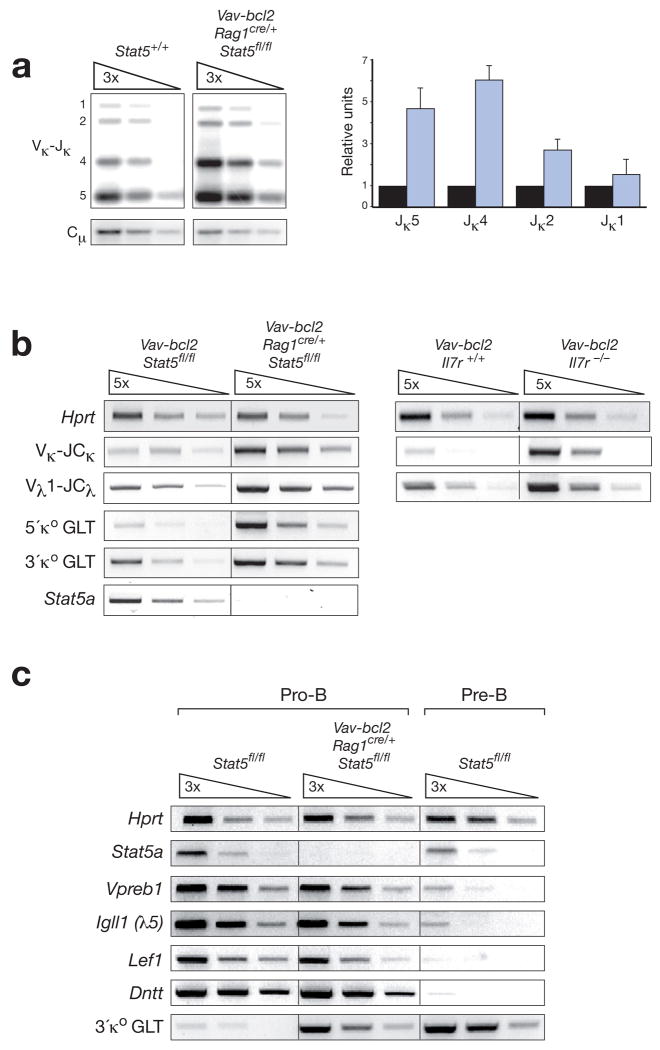
Figure 7. STAT5 and IL-7 signaling repress Igk and Igl gene rearrangements in pro-B cells.
(a) Increased Igk gene rearrangements in pro-B cells sorted from Vav-bcl2 Rag1cre/+ Stat5fl/fl mice (blue bar) compared to wild-type mice (black bar). Vκ gene rearrangements to the Jκ1 to Jκ5 gene segments (indicated by numbers along the left margin) were analyzed by semiquantitative PCR of 3-fold serially diluted DNA followed by Southern blot detection. The radioactive signals corresponding to these rearrangements were quantified by phosphorimager analysis, normalized to the value of the control Cμ PCR fragment and displayed in the bar graph to the right. The normalized value obtained for each Vκ-Jκ rearrangement in wild-type pro-B cells was set to 1. The quantification of 3 independent experiments is shown. (b) RT-PCR analysis of Igk transcripts. The expression of rearranged Vκ-JCκ and Vλ1-JCλ transcripts as well as the two κo germline transcripts (GLT) were analyzed by semiquantitative RT-PCR using 5-fold serial dilutions of cDNA prepared from FACS-sorted pro-B cells of the indicated genotypes. The 5’ and 3’ κo GLTs are transcribed from distinct promoters (see Supplementary Fig. 18a) and give rise to 1.1-kb and 0.8-kb spliced transcripts, respectively. (c) Expression of pro-B cell-specific genes in Bcl-2-rescued STAT5-deficient pro-B cells. FACS-sorted pro-B cells (c-Kit+CD19+CD25−IgM−) and pre-B cells (CD19+CD25+c-Kit−IgM−) of control Stat5+/+ or Vav-bcl2 Rag1cre/+ Stat5fl/fl mice were analyzed by semiquantitative RT-PCR for expression of the ‘pro-B cell-specific’ genes Vpreb1, Igll1 (λ5), Lef1 and Dntt (TdT)48 and the ‘pre-B cell-specific’ 3’ κo GLT.