Figure 1. Several lincRNAs are p53 transcriptional targets.
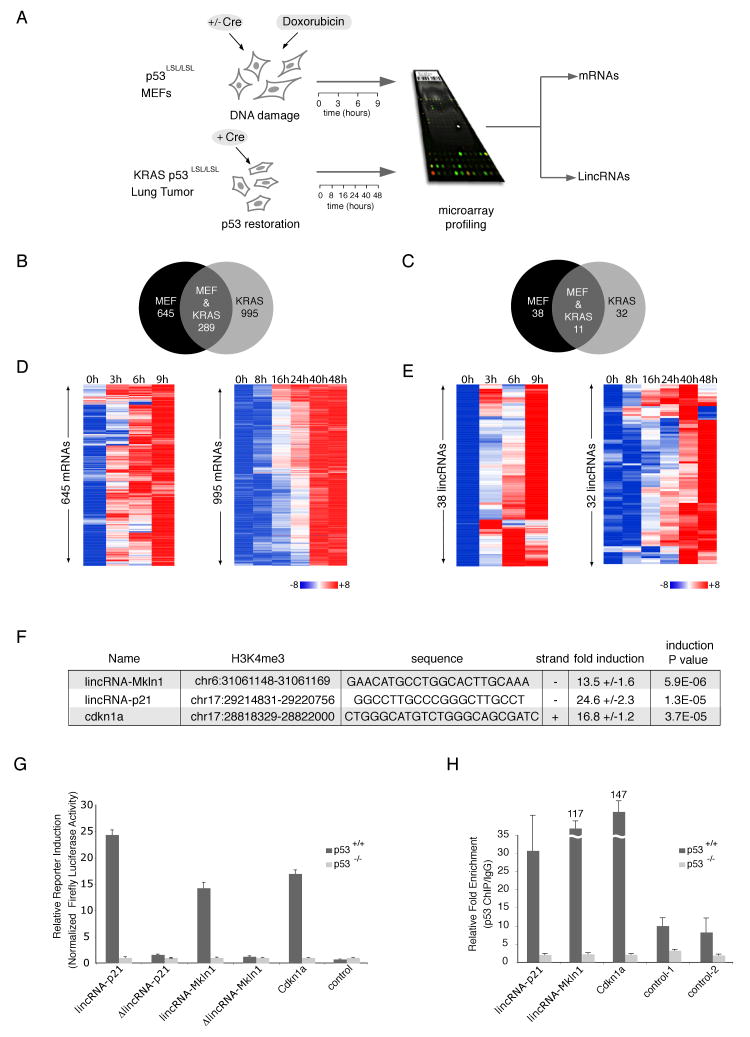
(A). Experiment layout to monitor p53-dependent transcription. p53-restored (+Cre) or not restored (-Cre) p53LSL/LSL MEFs were treated with 500nM dox for 0, 3, 6 and 9 hours (top left). KRAS (p53LSL/LSL) tumor cells were treated with hydroxytamoxifen for p53 restoration for 0, 8, 16, 24, 40 or 48 hours (bottom left). RNA was subjected to microarray analysis of mRNAs and lincRNAs.
(B) and (C). Venn diagrams showing the number of shared and distinct mRNAs (B) or lincRNAs (C) induced in a p53-dependent manner in the MEF or KRAS systems.
(D) and (E). mRNAs (D) and lincRNAs (E) activated by p53 induction (FDR < 0.05) in MEF or KRAS system. Colors represent transcripts above (red) or below (blue) the global median scaled to 8 fold activation or repression, respectively.
(F) Promoter region, conserved p53 binding motif, promoter orientation and p53-dependent fold induction in reporter assays of lincRNA promoters induced in a p53-dependent manner (values are average of at least three biological replicates (+/-STD); P values are determined by t-test)
(G) p53-dependent induction of lincRNA promoters requires the consensus p53 binding elements. Relative firefly luciferase expression driven by promoters with p53 consensus motif (lincRNA-p21, lincRNA-Mkln1) or with deleted motif (ΔlincRNA-p21 and ΔlincRNA-Mkln1) in p53-restored p53LSL/LSL (p53+/+) or p53LSL/LSL (p53-/-) cells. Values are relative to p53-/- and normalized by renilla levels (average of 3 replicates +/-STD).
(H) p53 specifically binds to p53 motifs in lincRNA promoters. p53 ChIP enrichment in p53+/+ and p53-/- MEFs on regions with p53 motifs (lincRNA-p21, lincRNA-Mkln1, Cdkn1a) or two irrelevant regions (controls). Enrichment values are relative to IgG and average of 3 replicates (+/-STD).
See also Figure S1 and Table S1.