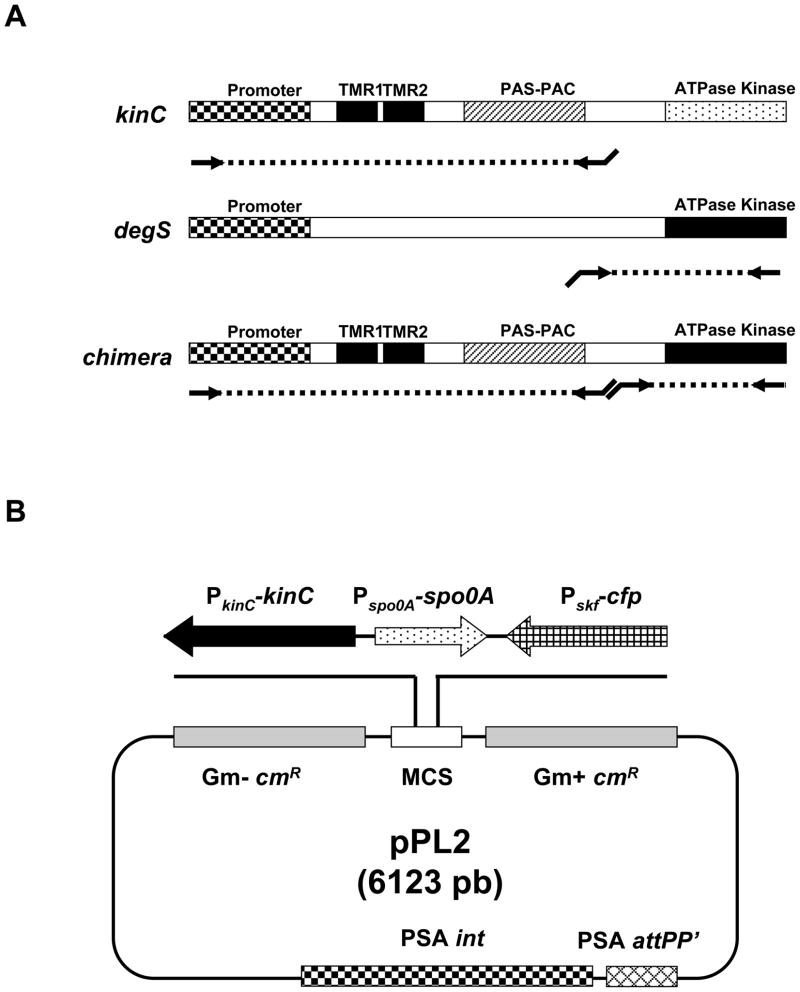
Figure 4.
(A) Scheme of joining PCRs used to generate the kinase chimera KinC-DegS. Both genes kinC and degS are represented as rectangles in which the coding regions of the domains are marked. PCR amplification of the regions of the genes required to generate the chimera are represented as dashed lines. Primers required for amplification are illustrated as arrows. The regions coding for the transmembrane domain and the PAS-PAC sensor domain of kinC was amplified and joined to the kinase domain of degS by the joining PCR as represented in the figure. (B) Construction of the cassette KinC-Spo0A-Pskf-cfp and integration into the genome of Listeria monocytogenes. The multicloning site (MCS) of the vector pPL2 was used to clone the joining PCR KinC-Spo0A-Pskf-cfp. This plasmid integrates into the genome of L. monocytogenes due to the PSA prophage integrase gene (PAS int) and the PSA prophage attachment site in tRNAArg (PSA attPP′). Selection is given by the chloramphenicol resistance marker (cmR). The plasmid harbors two different cmR genes in order to be selectable in gram-positive strains (Gm- cmR) and gram-negative strains (Gm+ cmR).