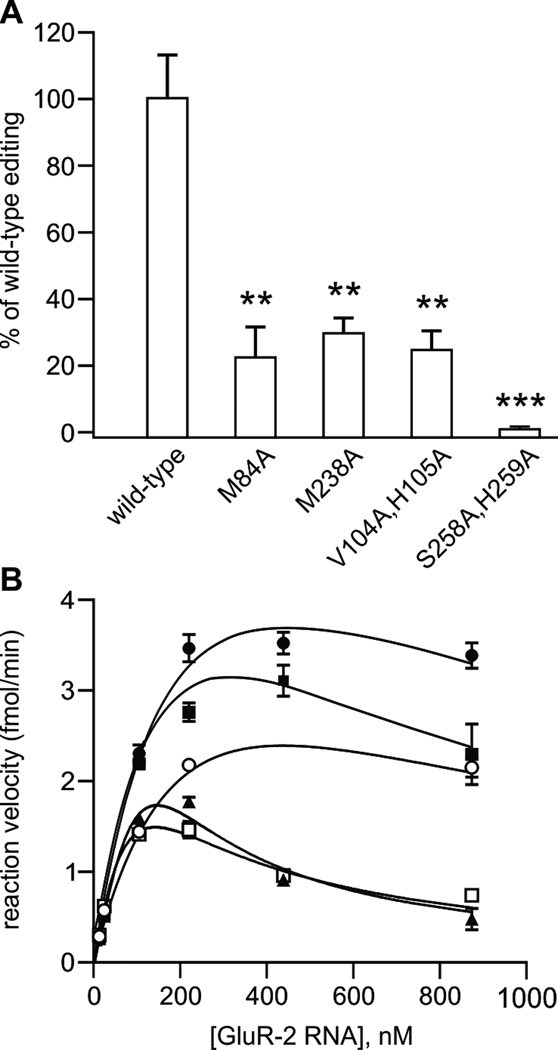
Figure 5.
Sequence-specific contacts of ADAR dsRBMs are important for editing activity. (A) Quantitative analysis of in vitro editing efficiency for ADAR2 dsRBM double mutants; all mutants were assayed in duplicate for in vitro editing activity at the GluR-2 R/G site using three independent nuclear extracts (mean ± SEM; *p<0.05, **p<0.005; ***p<0.001). (B) Kinetic analysis of wild-type ADAR2 editing with GluR-2 R/G mutants. Increasing concentrations of GluR-2 RNAs (see Figure S4; wild-type ●; mut 1 ■, mut 2 ○, mut 3 ▲, mut 4 □) were incubated with wild-type rat ADAR2 protein as described above; all mutant RNAs were assayed in triplicate for determination of in vitro reaction velocity (mean ± SEM). Non-linear fitting of kinetic curves corresponded to a model of substrate inhibition (R2 = 0.91–0.98 for all RNAs) with Vmax values corresponding to 3.92, 3.84, 2.08, 1.20 and 1.29 fmol/min for wild-type, mut1, mut2, mut3 and mut4, respectively. Wild-type and mutant sequences are shown in Figure S4.