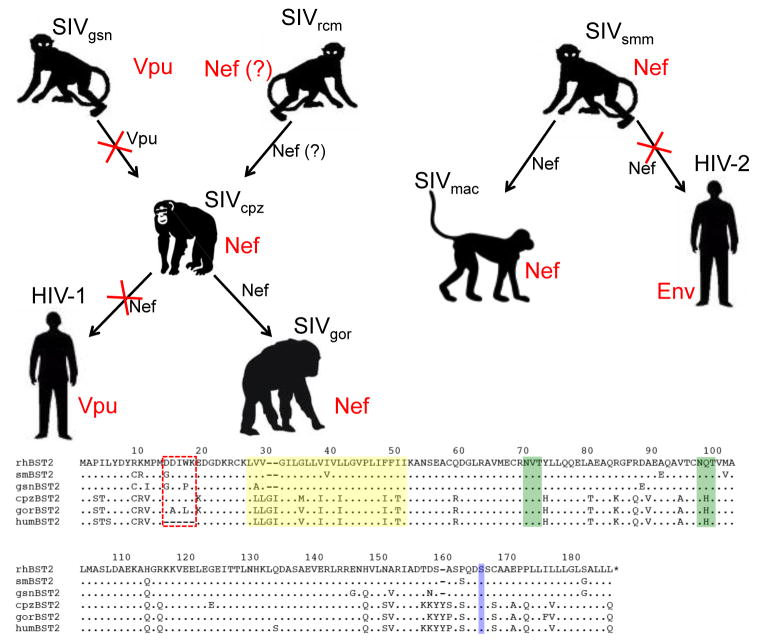
Figure 4. Adaptation of the primate lentiviruses for overcoming restriction by BST-2 in their respective hosts.
Chimpanzee SIV (SIVcpz) is thought to be a recombinant derived from Old World monkey SIVs similar to viruses presently found in the greater spot-nosed monkey (SIVgsn) and the red-capped mangabey (SIVrcm). HIV-1 and SIVgor are the result of the cross-species transmission of SIVcpz from chimpanzees to humans and from chimpanzees to gorillas. Likewise, HIV-2 and SIVmac reflect the cross-species transmission of SIVsmm from sooty mangabeys to humans and from sooty mangabeys to macaques. Arrows represent cross-species transmission events and the viral gene products that antagonize BST-2 in each species are indicated in red. The viral gene products that either retained, or lost (indicated by red X’s). Activity against BST-2 are indicated in black next to each arrow. Species-specific differences in BST-2 are depicted in the amino acid sequence alignment. Representative BST-2 sequences are shown for the rhesus macaque (rhBST2), sooty mangabey (smBST2), greater spot-nosed monkey (gsnBST2), chimpanzee (cpzBST2), gorilla (gorBST2) and humans (humBST2). Residues of the cytoplasmic domain that confer susceptibility to Nef are indicated by the red box, the transmembrane domain is highlighted in yellow, potential N-linked glycosylation sites are indicted in green and the site for cleavage and addition of the GPI anchor is indicated in blue. The five-residue deletion in the cytoplasmic domain of human BST-2 renders the protein resistant to antagonism by Nef, leading to the acquisition of BST-2 antagonist activity by HIV-1 Vpu and HIV-2 Env.