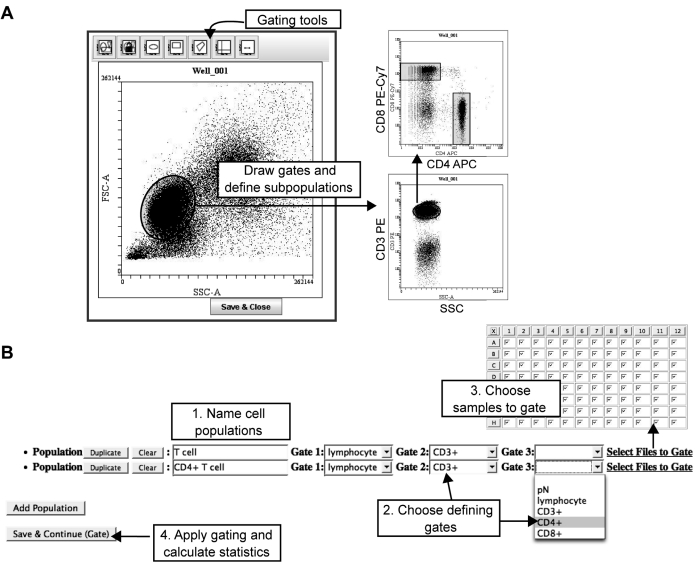
FIG. 3.
Definition of cell populations in WebFlow. Screenshots of WebFlow gating and populations definition pages. (A) Gating cell subsets. The gating applet of WebFlow provides multiple gating tools including ellipses, rectangles, polygon, quadrant, and histogram gates. The user can draw gates, gate the sample, and then define further subgates. Here, the user gated lymphocytes based on forward and side light scatter (FSC and SSC), then gated on CD3+ T cells. The CD4+ and CD8+ T cell subsets were gated based on CD4 and CD8 levels. (B) Population definition. Once gates have been drawn, the user names cell populations (Step 1) and chooses the gates that define that population (Step 2). The user then selects which samples to apply the population analysis to (Step 3), allowing different analyses to be applied to different plates. Once all populations have been defined, the final step instructs WebFlow to calculate standard statistics (mean, median, CV, percentage, number) for each parameter on each defined cell population (Step 4). In this example, CD4+ T cells are defined by the lymphocyte, CD3+, and CD4+ gates. All samples in the plate were stained the same way, so the entire plate is selected for analysis.