Abstract
DNA damage tolerance pathways facilitate the bypass of DNA lesions encountered during replication. These pathways can be mechanistically divided into recombinational damage avoidance and translesion synthesis, in which the lesion is directly bypassed by specialised DNA polymerases. We have recently shown distinct genetic dependencies for lesion bypass at and behind the replication fork in the avian cell line DT40, bypass at the fork requiring REV1 and bypass at post-replicative gaps requiring PCNA ubiquitination by RAD18. The WRN helicase/exonuclease, which is mutated in the progeroid and cancer predisposition disorder Werner's Syndrome, has previously been implicated in a RAD18-dependent DNA damage tolerance pathway. However, WRN has also been shown to be required to maintain normal replication fork progression on a damaged DNA template, a defect reminiscent of REV1-deficient cells. Here we use the avian cell line DT40 to demonstrate that WRN assists REV1-dependent translesion synthesis at the replication fork and that PCNA ubiquitination-dependent post-replicative lesion bypass provides an important backup mechanism for damage tolerance in the absence of WRN protein.
Keywords: DNA damage tolerance, Translesion synthesis, Werner's Syndrome, PCNA ubiquitination, WRN, REV1
1. Introduction
During DNA replication, attempts to excise and repair DNA damage are likely to result in replication fork collapse. Cells therefore usually opt to bypass damage encountered by replication forks, instead deferring repair until replication has been completed. DNA damage bypass can take two general forms [for recent reviews see [1,2]]. The most direct is translesion synthesis, in which the stalled replicative polymerases are replaced by specialised translesion polymerases whose active sites are able to accommodate adducted or distorted bases. Alternatively, cells can employ one of the recombinational modes of bypass that require an undamaged template, usually the newly synthesised daughter strand on the sister chromatid. In principle, lesion bypass can take place either at the replication fork or at post-replicative gaps created when replication restarts downstream of an arrested fork. We have recently demonstrated that, in DT40, these temporally distinct lesion bypass pathways are genetically defined by requirements for the C terminus of the Y-family DNA polymerase REV1 and the monoubiquitination of the DNA sliding clamp PCNA respectively [3]. Monoubiquitination of PCNA by the E3 ubiquitin ligase RAD18 [4–6] plays a central role in the control of DNA damage tolerance in all eukaryotes studied to date [4–8] both through controlling the recruitment of translesion polymerases and by promoting recombinational bypass. However, while more is now understood about the mechanisms that control lesion bypass pathways, little is known about the accessory factors that facilitate these processes. A number of recent lines of evidence, discussed below, suggest that WRN is potentially one such factor.
Werner's Syndrome (WS, OMIM 277700) is an autosomal recessive disorder caused by mutations in the WRN gene [9] and is characterised by a variety of disorders reminiscent of premature ageing; cataracts, type II diabetes mellitus, osteoporosis, atherosclerosis as well as a high incidence of unusual cancers. WRN encodes a member of the RecQ family of helicases [10] but also contains a ‘helicase, RNaseD, C-terminal conserved’ (HRDC) domain, which has DNA-binding activity [11,12], and 3′–5′ exonuclease activity due to a motif located near the N terminal of the protein [13].
The defects in WRN-deficient cells, which include slow growth, spontaneous genetic instability and sensitivity to a range of DNA damaging agents, arise from the involvement of WRN in a very wide range of DNA transactions including base excision repair, non-homologous end joining and transcription [reviewed in [14]]. Much evidence, however, points to its involvement in processes at or around the replication fork. Similarly to RecQ in E. coli [15] and Rqh1 in S. pombe [16], there is evidence that WRN helps to avoid recombination [17], possibly by promoting damage tolerance, the set of mechanisms that promote DNA damage bypass during replication. WRN has also been reported to play a role in homologous recombination itself at the level of the resolution of recombination intermediates [18–21].
Evidence implicating WRN in DNA damage tolerance pathways comes from a number of sources. WRN interacts functionally with the Y-family translesion polymerases η, κ and ι, stimulating their extension activity in the absence of PCNA [22]. WRN alleviates pausing of these polymerases at lesions by increasing the vmax of polymerization, hence it was proposed that it may promote the progression of replication forks at the expense of increased mutagenesis. This stimulatory effect of WRN on TLS polymerases was found to be present even in WRN mutants lacking either helicase or exonuclease activities. WRN also co-localises with pol η after UV-C irradiation, but formation of WRN foci did not depend on the presence of pol η [22] and a direct interaction between the proteins has not been shown.
Genetic studies in DT40 suggest that WRN functions in a RAD18-dependent DNA damage tolerance pathway [23] with a wrn/rad18 double mutant exhibiting sensitivity to NQO or MMS equal to that of the rad18 single mutant. From our previous work showing that RAD18 defines a post-replicative pathway of damage bypass [3], these data suggest that WRN too would act behind the replication fork. However, using DNA fibre labelling, WRN has been shown to be required for normal replication fork progression on DNA damaged with MMS [24], suggestive of a model in which WRN assists bypass of lesions at the replication fork. This phenotype is reminiscent of REV1-deficient cells in similar fibre labelling assays [3,25] and suggests that WRN is required at the replication fork.
To resolve the question of which of the temporally distinct bypass pathways, defined by RAD18-dependent PCNA ubiquitination and by REV1 [3], WRN is involved in we generated isogenic mutants in the chicken cell line DT40. Using a newly created wrn DT40 that lacks any detectable transcript, we show that WRN operates in a pathway dependent on REV1 to maintain replication fork progression following DNA damage and that RAD18-dependent post-replicative gap filling provides an important backup activity when WRN is lost.
2. Materials and methods
2.1. DT40 lines, culture and transfection
Culture and transfection of DT40 was carried out as described previously [26]. The rev1 and pcnaK164R cell lines have also been previously described [26,27]. The rad18 mutants used were generated in our laboratory using constructs described by Takeda and co-workers [28].
2.2. WRN locus targeting construct assembly
The WRN targeting construct was assembled in pBluescript SK+ using a custom multiple cloning site (KpnI-NotI-MluI-ApaI-EcoRI-BamHI-SacI) made by annealing and ligating the oligonucleotides cWRNMCSF [5′-CGCGGCCGCACGCGTGGGCCCGAATTCGGATCCGAGCT] and cWRNMCSR [5′-CGGATCCGAATTCGGGCCCACGCGTGCGGCCGCGGTAC]. The 2.89 kb 5′ arm of homology was amplified by PCR from DT40 genomic DNA using primer pair cWRKO5F2 [5′-CAGAGAGAATAACGCGTCATAAAGATTGTATCTAAATTTCTAGTCTTC] and cWRKO5R2 [5′-CTCAGGAAACAGCCACATACAAAAGGGCCCGTAATAGTTTCCAGTTCC], which introduce MluI and ApaI sites at the 5′ and 3′ ends respectively. The 5.54 kb 3′ arm of homology was amplified from genomic DNA using primers cWRKO3F1 [5′-GGTGTTTGTTTGCCTGTGGCCGGGCATGG] and cWRKO3R1 [5′-CCCATAAGCTCTGAGCTCTTGGGCCATGTTG] which introduce a BamHI site at the 5′ end of the arm. An endogenous SacI site is present at the 3′ end. The 5′ arm was inserted into the modified pBluescript first, followed by the 3′ arm and then an antibiotic resistance cassette (either neomycin, puromycin, blasticidin or histidinol) was introduced into the BamHI site. The neomycin, puromycin and blasticidin resistance markers are removable using Cre recombinase as they are flanked by modified LoxP sites [29].
2.3. Confirmation of WRN disruption
After targeting of the first and second alleles, disruption was confirmed by Southern blot probing from the 3′ end (Fig. 1A and B). Probe DNA for Southern blotting was made by amplification of DT40 genomic DNA using the primers cWRKOPF1 [5′-GTTAATACCGTGGCTTGCTGAAGCATTTCTTGAC] and cWRKOPR2 [5′-GCTCAGCTGTAGGCTTTTGTTTTAATGAACACAAC], cloned into pCR 2.1-TOPO then excised as a SpeI, XhoI fragment. Genomic DNA from transfected clones was digested with BamHI before Southern blotting.
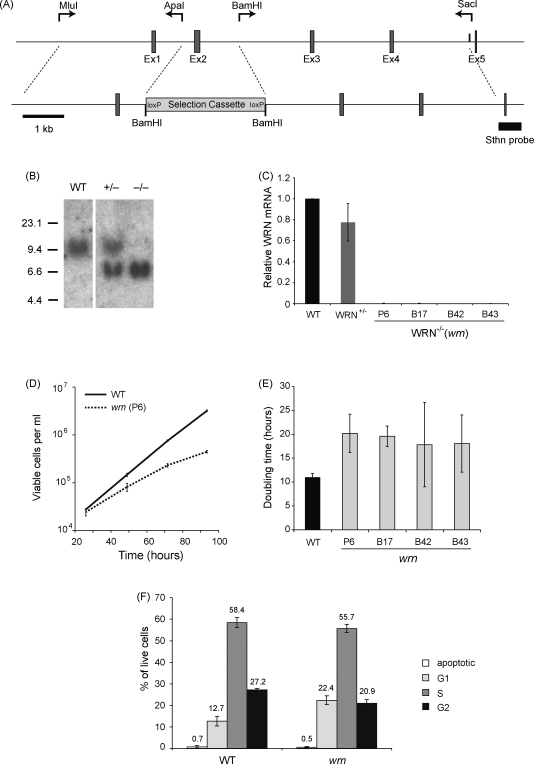
Fig. 1.
Generation of a WRN null DT40 line. (A) Schematic of the WRN gene targeting strategy. The binding sites of the primers used to generate the 5′ and 3′ arms of the targeting construct are shown above the locus map, along with the restriction sites used in their cloning. A selection cassette replaces exon 2 of the WRN transcript and which, following targeted integration, is predicted to lead to an out of frame splice event between exons 1 and 3 resulting in a stop codon 16 amino acids into exon 3. The Southern probe spanning exon 5 is indicated. (B) BamHI digested genomic DNA from wild type (WT), WRN+/−and WRN−/−(wrn) cells probed as indicated above. All lanes are from the same blot, but intervening lanes have been cut out between WT and +/−. (C) qPCR for exons 19–23 confirming loss of WRN transcript downstream of the disruption in four clones. (D) Growth of wild type (WT) and wrn clone P6. The graph represents three independent experiments with the error bars indicating the standard deviation. (E) Comparison of the doubling time in four independent clones of wrn DT40. (F) Cell cycle distribution of cells in asynchronous cultures of wild type and wrn lines. The mean percentage of 3 independent determinations is shown and the error bars represent one standard deviation.
Loss of WRN transcript was also confirmed by qPCR, carried out using a 7900HT Fast Real-Time PCR System (Applied Biosystems). Reactions were performed in MicroAmp fast optical 96-well reaction plate (Applied Biosystems) sealed with MicroAmp Optical Adhesive Film (Applied Biosystems) using SYBR GreenER qPCR SuperMix Universal (Invitrogen) and ROX reference dye. All reactions were carried out using cDNA template with primers at 400 μM. Primers to amplify WRN cDNA were designed to hybridize to the exons 19–20 and exons 22–23 junctions (Exon1920F has sequence 5′-GCGGGATGAAATCCAGTGTGTTGTGG and Exon2322R has sequence 5′-GAGGTTGCCAAACAAGAGGTTCTGCACCTG) Relative amounts of transcripts were determined using comparative quantitation relative to wild type, using β-actin as the normaliser. Primers used to amplify β-actin were BActinF1 [5′-GAGAGAAGATGACACAGATCATG] and BActinR1 [5′-GACTCCATACCCAAGAAAGATGG].
2.4. Measurement of growth kinetics
Cells were diluted to a density of 1 × 104/ml in medium and incubated (37 °C). Viable cells were counted using a Vi-CELL cell viability analyser (Beckman Coulter) at 24 h intervals for 96 h.
2.5. Cell cycle analysis
Cell cycle analysis was performed as previously described [30]. Incorporated 5-bromo-2′deoxyuridine was stained using a 1:200 dilution of rat anti-BrdU antibody (BD Biosciences) followed by a 1:100 dilution of FITC-conjugated goat anti-rat antibody (BD Biosciences). FACS analysis was carried out using a FACScalibur flow cytometer (Beckman Coulter) and FlowJo 8.5.2 software (Tree Star/Stanford University).
2.6. Post-replication repair assay
Post-replication repair assays were carried out as previously described [3].
2.7. Colony survival assays
Colony survival assays were performed as previously described [31].
2.8. DNA fibre spreading and labelling
DNA fibre spreading and labelling was carried out as described [3]. 100 doubly labelled fibres were counted for each condition. These were derived from c. 8 slides made from two independent labelling experiments.
3. Results and discussion
3.1. Generation of a completely null wrn DT40 line
For this study we wished to produce a completely null wrn DT40 cell line, since an earlier wrn DT40 mutant retains a substantial transcript (2.5 of 4.5 kb) as assessed by Northern blot [32]. To achieve this we replaced exon 2 in both alleles of the DT40 WRN gene with a selectable marker by homologous recombination (Fig. 1A and B). Splicing of exon 1 to exon 3 results in an out of frame transcript and introduces a stop codon 16 amino acids into exon 3. qPCR corresponding to exons 19–23 on cDNA from the targeted clones confirmed the success of this strategy with no transcript detectable (Fig. 1C).
3.2. The phenotype of wrn DT40 resembles that of WS patient cells
Our wrn DT40 clones grew slowly, as has been reported for human WS fibroblast lines [33]. We initially characterised four independent wrn clones, all of which showed a comparable reduction in doubling time. For example clone P6 had doubling time at 37 °C of 20.2 ± 4.0 h compared to 11.0 ± 0.8 h for the parental wild type, measured concurrently (Fig. 1D and E). Similar to human WS fibroblasts [reviewed in [34]], this was largely explained by a prolonged cell cycle rather than an increase in spontaneous apoptosis (Fig. 1F). The increase in the proportion of G1/G0 cells in an asynchronous culture of the wrn mutant to 22.7%, from 12.7% in wild type, corresponds to wrn cells spending an average of 4.5 h in G1 as opposed to 1.4 h for wild type (Fig. 1F). Further, S-phase was also prolonged from 6.4 h in wild type to 11.3 h in wrn cells, again in agreement with studies of human WS cells [33,35,36]. Similar to human WS fibroblasts [37–42], wrn DT40 were modestly sensitive to a number of agents expected to create replication blocks including ultraviolet light [D10 8.3 vs. 4.8 J/m2, for wild type (Fig. 2A)], the adduct/cross-linking agent cisplatin (CDDP) [D10 34.1 vs. 45.7 μM (Fig. 2A)], methylmethane sulphonate (MMS) [D10 99.8 vs. 142.1 parts per million (data not shown)] and nitroquinoline-1-oxide (NQO) [D10 23.7 vs. 41.5 nM (data not shown)].
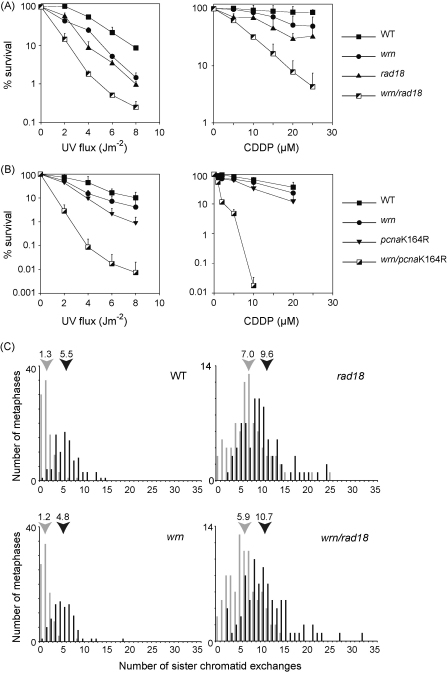
Fig. 2.
WRN and PCNA ubiquitination act in separate pathways. (A) Epistasis analysis of WRN and RAD18 for survival following treatment with 254 nm UV light (UV) and cisplatin (CDDP). (B) Epistasis analysis of WRN and cells carrying a point mutation of PCNA at K164, which prevents ubiquitination, for survival following treatment with 254 nm UV light (UV) and cisplatin (CDDP). Error bars represent the one standard deviation from the mean of three independent experiments. For clarity only the positive error bar is shown. C. Sister chromatid exchange in wrn and rad18 cells. SCE with (black bars) and without (light grey bars) treatment with 0.2 ng/ml NQO. The histogram indicates the percentage of metaphases with the number of SCE indicated on the X-axis. The mean number of SCE in each case, derived from blind scoring of at least 100 metaphases, is indicated above the histogram. For both untreated and NQO-treated cases, the difference between wild type and rad18 SCE is statistically significant (unpaired t-test, p < 0.0001). There is no significant difference between wild type and wrn or rad18 and wrn/rad18.
3.3. Complete WRN disruption is not epistatic with defective PCNA ubiquitination
Previous work has suggested that WRN functions in a RAD18-dependent damage avoidance pathway [23]. We re-examined this relationship by disrupting the WRN loci using our construct in rad18 [28] and pcnaK164R [27] DT40 lines. Epistasis analysis for sensitivity to 254 nm UV light and cisplatin (CDDP) showed that both wrn/rad18 and wrn/pcnaK164R double mutants were markedly more sensitive than either of their respective single mutants (Fig. 2A and B), showing that WRN acts largely independently of PCNA ubiquitination and, indeed, suggests that PCNA ubiquitination plays an important role in maintaining the viability of wrn cells following genotoxic stress.
A striking feature of rad18 mutant lines is elevated spontaneous sister chromatid exchange [28,43]. This arises from a combination of channelling DNA lesions away from RAD18-dependent DNA damage tolerance pathways to classical homologous recombination and from a role played by RAD18 in recombination itself [44] that is independent of PCNA ubiquitination [6]. In contrast, it has been reported that inactivation of WRN in human cells does not result in elevated SCE [45]. As previously observed, rad18 DT40 cells exhibit elevated levels of SCE in the absence of any exogenously applied DNA damage, with a mean of 7 per metaphase (Fig. 2C). The level of SCE rose further following treatment with NQO to 10 per metaphase (Fig. 2C). However, in agreement with studies in human WS cells [45], wrn DT40 do not exhibit elevated spontaneous SCE or an exaggerated SCE response following exposure to NQO and the wrn/rad18 double mutant behaves the same as the rad18 single mutant within the sensitivity limit of this assay (Fig. 2B).
3.4. WRN is not required for post-replicative gap filling
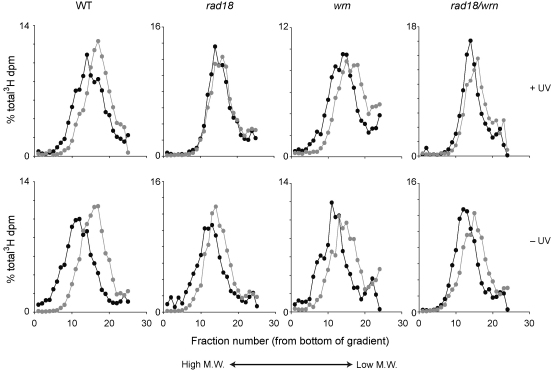
In DT40, RAD18-mediated PCNA ubiquitination controls DNA damage bypass at post-replicative gaps that are formed in consequence of replication fork arrest with downstream resumption of replication [3]. Thus, when PCNA ubiquitination is defective the rate at which single stranded gaps are filled is reduced, i.e. PCNA ubiquitination mutants exhibit defective post-replication repair. A key prediction of the notion that WRN functions in a RAD18-dependent damage avoidance pathway [23] is that WRN should also operate behind the fork. This can be monitored by following the incorporation of a tract of tritiated label into high molecular weight single stranded DNA by velocity sedimentation on an alkaline sucrose gradient [46]. We observed, as previously reported [3,47] that rad18 cells exhibit delayed gap filling (Fig. 3). However, such a defect is not seen in wrn cells and the defect in the wrn/rad18 line is comparable to that seen in the rad18 single mutant (Fig. 3). This suggests that WRN is not essential for timely post-replicative gap filling and together these data do not support WRN acting in the PCNA ubiquitination-dependent post-replicative DNA damage tolerance pathway.
Fig. 3.
WRN is not required for post-replicative gap filling. Upper panels: Wild type, rad18, wrn and rad18/wrn cells were irradiated with 4 J m−2 254 nm light, pulse labelled with [3H]-thymidine for 20 min and either lysed immediately (grey lines) or chased for 90’ in medium containing 10 μM thymidine before lysis (black lines). Lower panels: sham-irradiated cells.
Our conclusions are, on the face of it, in marked contrast to those reached by Dong et al. [23]. We believe that the discrepancy likely arises from the incomplete inactivation of WRN in the DT40 mutant used in the earlier study [32]. Although the targeting approached used in the work of Dong et al. will inactivate the helicase domain, most cases of WS are a result of complete functional inactivation of the gene. Loss of just the helicase activity of WRN results has been previously shown to result in a distinct and milder phenotype compared with complete inactivation of the protein [21]. Thus, while we cannot exclude that the helicase activity of WRN participates in PCNA ubiquitination-dependent damage avoidance, our results clearly demonstrate that the complete WRN protein fulfils an important function in the absence of PCNA ubiquitination. To further address this issue, we have tried extensively to perform complementation of our wrn mutant. However, we have been unable to obtain clones ectopically expressing the WRN protein despite using a number of different promoter systems. It will therefore be necessary to further these studies by creating endogenous mutations in the WRN locus.
3.5. WRN is required to maintain replication fork progression on damaged DNA
Recent data from WRN-depleted human cells have implicated WRN in maintaining replication fork progression on a damaged DNA template [24]. Fork progression is monitored by labelling DNA in vivo with halogenated nucleotides and then spreading the DNA fibres on a glass slide. The tracts of labelled DNA are revealed by denaturation and staining with antibodies that recognise halogenated bases. In our version of this assay [3], we sequentially labelled cells with chlorodeoxyuridine (CldU) and iododeoxyuridine (IdU). During the second labelling period we damaged the DNA such that the advancing fork will encounter DNA damage and may stall (Fig. 4A). We can then monitor, in a large number of fibres, the length of tract replicated before and after the application of DNA damage.
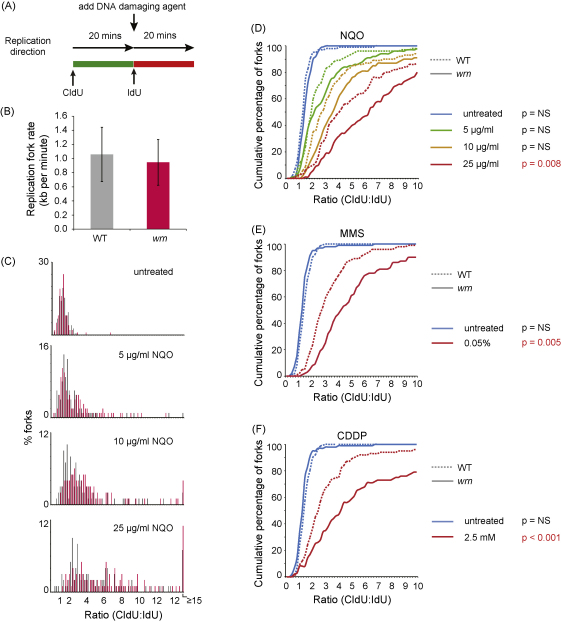
Fig. 4.
WRN is required to maintain replication fork progression on a damaged DNA template. (A) Schematic of DNA labelling experiments. Cells are pulse labelled first with chlorodeoxyuridine (CldU) for 20 min and then with iododeoxyuridine (IdU) for 20 min. DNA damage can be added along with the IdU to assess the effect on replication progression compared with the first labelling period without damage. Each period of labelling corresponds to synthesis of about 20 kb of DNA. Note the concentrations of drug used are supralethal in order to achieve a high probability of damage forming within c. 20 kb ahead of the fork [3]. (B) Fork progression on undamaged DNA in wrn cells is comparable to wild type (WT). Average fork rate during the first 20 min calculated using a previous calibration for this method [51] from 100 doubly labelled fibres for each case. The error bars indicate one standard deviation. There is no significant difference between WT and wrn (unpaired t-test). (C) Histograms of DNA fibre CldU:IdU ratios showing replication stalling in response to increasing doses of 4-nitroquinoline-1-oxide (NQO). Wild type: grey bars; wrn: red bars. (D–F) Cumulative percentage plots of DNA fibre CldU:IdU ratios. WT: dashed lines; wrn: solid lines. (D) 4-nitroquinoline-1-oxide (NQO). (E) Methylmethane sulphonate (MMS). (F) Cisplatin (CDDP). Statistical significance was assessed using the two-sample Kolmogorov–Smirnov (K–S) test and the probability that there is no difference between WT and wrn at each dose is indicated.
In the absence of DNA damage the replication rate of wrn cells was comparable to wild type at around 1 kb/min (Fig. 4B). To assess the effect of disruption of WRN on fork progression following DNA damage we performed titrations in which increasing doses of DNA damaging agent were added during the second labelling period. Damage arresting replication during the second labelling period will result in shortening of the IdU labelled tract and an increase in the ratio of CldU:IdU tract lengths. As the position of the stall is random, this ratio increases stochastically resulting in both an increase in the mean CldU:IdU ratio and in the spread of values (Fig. 4C). A convenient method to represent this data is as a cumulative percentage of forks at each ratio (Fig. 4D–F). For all doses of NQO, methyl methane sulphonate and cisplatin wrn cells (solid lines) exhibited a more profound inability to maintain replication fork progression on a damaged template compared with wild type (dashed lines), in agreement with the recent work on human cells [24].
3.6. WRN functions at the replication fork to facilitate REV1-dependent translesion synthesis
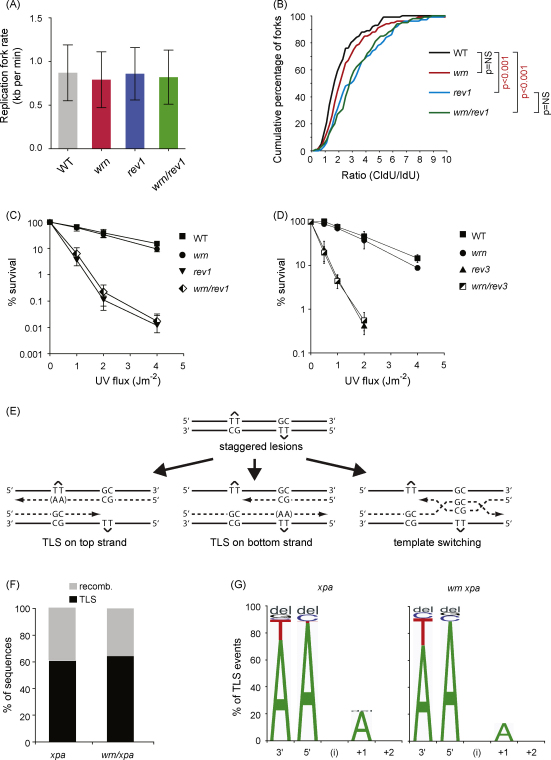
This phenotype is strongly reminiscent of cells lacking REV1 [3,25]. REV1 plays a crucial role in coordinating translesion synthesis at the replication fork by coordinating, through its C terminus, interactions between PCNA and translesion polymerases [3,48,49]. We therefore wished to know whether this phenotypic similarity between WRN and REV1 was a reflection of their operating in common damage bypass pathway at the replication fork. To do this we created a wrn/rev1 double mutant. In the absence of damage these cells show no defect in DNA replication rate on undamaged DNA (Fig. 5A). To examine the effect of DNA damage we chose a dose of NQO (10 μg/ml) at which we would expect to be able to readily detect an additive effect of the double disruption (see Fig. 4D). In a side-by-side comparison, rev1 cells exhibited a more profound defect in fork progression following damage than wrn cells (Fig. 4B), but importantly the double wrn/rev1 mutant was no worse than rev1 on its own. This suggests that WRN operates at a subset of replication impediments at the fork that require REV1 for their bypass or resolution.
Fig. 5.
WRN acts in a REV1-dependent pathway to promote replication fork progression on damaged DNA. (A) Fork progression on undamaged DNA in wild type (WT), wrn, rev1 and wrn/rev1 cells. There is no significant difference between any of the lines (unpaired t-test). (B) Cumulative percentage plots of DNA fibre CldU:IdU ratios with 10 μg/ml NQO added during the second (IdU) labelling period for wild type (black line), wrn (red line), rev1 (blue line) and wrn/rev1 green line. Statistical significance assessed by the two-sample K-S test is indicated. (C and D) Epistasis analysis of WRN and REV1 (C) and REV3 (D) for survival following treatment with 254 nm UV light. E. Summary of the replicating plasmid assay for lesion bypass. For full details see Szüts et al. [50]. Briefly, the plasmid pQTS, which can support replication in DT40, contains staggered T–T (6-4) photoproducts placed opposite a GpC mismatch, since this is an uncommon insertion opposite this lesion. This arrangement allows determination of TLS and recombinational bypass by template switching. Since GpC opposite the lesion would also arise as a result of nucleotide excision repair, the assay is carried out in an xpa background. (F) The proportion of TLS vs. error-free bypass in pQTs sequences recovered from xpa (n = 130) and xpa/wrn cells (n = 70), shown as percentage of the total. (G) The pattern of nucleotide incorporation opposite the T–T (6-4) photoproduct. The percentage of each nucleotide incorporated at each position is indicated by the size of the letter of the nucleotide in the column; del: deletion.
We next examined the genetic relationship between WRN and REV1 and REV3 for sensitivity to UV light. These experiments revealed a clear epistatic relationship between WRN and REV1/3-dependent translesion synthesis, the double mutants exhibiting no additional sensitivity to the single TLS mutants (Fig. 5C and D). This is in marked contrast to the relationship between WRN and PCNA ubiquitination (Fig. 2B). Together, these data suggest that WRN and REV1 can operate in a common pathway for the alleviation of replication arrest.
We have previously shown that REV3 is essential for translesion synthesis across (6-4) T–T photoproducts, a major UV-induced DNA lesion, in vivo in a replicating plasmid assay that can monitor the relative use of TLS and recombinational bypass (Fig. 5E) [50]. We also showed that loss of REV1 not only diminished the frequency of use of TLS but also resulted in reduced TLS frame fidelity. To examine the impact of the wrn mutation in this assay, we inactivated nucleotide excision repair, as previously described, by disrupting the XPA locus [50]. Since the lesion is placed opposite a G–C mismatch, this allows the distinction of recombinational bypass from excision repair. Interestingly, wrn/xpa cells exhibited a very similar frequency of TLS usage (Fig. 5E) and no alteration in the mutation spectrum created by the use of TLS compared with xpa cells (Fig. 5F). This suggests that WRN does not directly influence the TLS reaction itself or impact mutagenesis, at least in this context. Rather it appears to be required for efficient use of TLS at a subset of events at the fork. It will ultimately be interesting to determine the circumstances under which WRN is required. Nonetheless, in the absence of WRN, we suggest that bypass is more likely to take place behind the fork explaining the marked reliance of wrn mutant cells on PCNA ubiquitination.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Acknowledgements
Work in our lab is supported by the Medical Research Council and Association for International Cancer Research. We would like to thank Shunichi Takeda and Jean-Marie Buerstedde for sharing DT40 mutants and targeting constructs, Shige Iwai for preparing the T–T(6-4) photoproduct containing oligonucleotides, Dávid Szüts for assembling the replicating plasmid and members of the group for discussions.
References
- 1.Andersen P.L., Xu F., Xiao W. Eukaryotic DNA damage tolerance and translesion synthesis through covalent modifications of PCNA. Cell Res. 2008;18:162–173. doi: 10.1038/cr.2007.114. [DOI] [PubMed] [Google Scholar]
- 2.Budzowska M., Kanaar R. Mechanisms of dealing with DNA damage-induced replication problems. Cell Biochem. Biophys. 2009;53:17–31. doi: 10.1007/s12013-008-9039-y. [DOI] [PubMed] [Google Scholar]
- 3.Edmunds C.E., Simpson L.J., Sale J.E. PCNA Ubiquitination and REV1 define temporally distinct mechanisms for controlling translesion synthesis in the avian cell line DT40. Mol. Cell. 2008;30:519–529. doi: 10.1016/j.molcel.2008.03.024. [DOI] [PubMed] [Google Scholar]
- 4.Hoege C., Pfander B., Moldovan G.L., Pyrowolakis G., Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 5.Kannouche P.L., Wing J., Lehmann A.R. Interaction of human DNA polymerase η with monoubiquitinated PCNA: a possible mechanism for the polymerase switch in response to DNA damage. Mol. Cell. 2004;14:491–500. doi: 10.1016/s1097-2765(04)00259-x. [DOI] [PubMed] [Google Scholar]
- 6.Simpson L.J., Ross A.L., Szuts D., Alviani C.A., Oestergaard V.H., Patel K.J., Sale J.E. RAD18-independent ubiquitination of proliferating-cell nuclear antigen in the avian cell line DT40. EMBO Rep. 2006;7:927–932. doi: 10.1038/sj.embor.7400777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frampton J., Irmisch A., Green C.M., Neiss A., Trickey M., Ulrich H.D., Furuya K., Watts F.Z., Carr A.M., Lehmann A.R. Postreplication repair and PCNA Modification in Schizosaccharomyces pombe. Mol. Biol. Cell. 2006 doi: 10.1091/mbc.E05-11-1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leach C.A., Michael W.M. Ubiquitin/SUMO modification of PCNA promotes replication fork progression in Xenopus laevis egg extracts. J. Cell Biol. 2005;171:947–954. doi: 10.1083/jcb.200508100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu C.E., Oshima J., Fu Y.H., Wijsman E.M., Hisama F., Alisch R., Matthews S., Nakura J., Miki T., Ouais S., Martin G.M., Mulligan J., Schellenberg G.D. Positional cloning of the Werner's syndrome gene. Science. 1996;272:258–262. doi: 10.1126/science.272.5259.258. [DOI] [PubMed] [Google Scholar]
- 10.Gray M.D., Shen J.C., Kamath-Loeb A.S., Blank A., Sopher B.L., Martin G.M., Oshima J., Loeb L.A. The Werner syndrome protein is a DNA helicase. Nat. Genet. 1997;17:100–103. doi: 10.1038/ng0997-100. [DOI] [PubMed] [Google Scholar]
- 11.Morozov V., Mushegian A.R., Koonin E.V., Bork P. A putative nucleic acid-binding domain in Bloom's and Werner's syndrome helicases. Trends Biochem. Sci. 1997;22:417–418. doi: 10.1016/s0968-0004(97)01128-6. [DOI] [PubMed] [Google Scholar]
- 12.von Kobbe C., Bohr V.A. A nucleolar targeting sequence in the Werner syndrome protein resides within residues 949–1092. J. Cell Sci. 2002;115:3901–3907. doi: 10.1242/jcs.00076. [DOI] [PubMed] [Google Scholar]
- 13.Huang S., Li B., Gray M.D., Oshima J., Mian I.S., Campisi J. The premature ageing syndrome protein, WRN, is a 3’-- > 5’ exonuclease. Nat. Genet. 1998;20:114–116. doi: 10.1038/2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rossi M.L., Ghosh A.K., Bohr V.A. Roles of Werner syndrome protein in protection of genome integrity. DNA Repair (Amst.) 2010;9:331–344. doi: 10.1016/j.dnarep.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hanada K., Ukita T., Kohno Y., Saito K., Kato J., Ikeda H. RecQ DNA helicase is a suppressor of illegitimate recombination in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 1997;94:3860–3865. doi: 10.1073/pnas.94.8.3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stewart E., Chapman C., Al-Khodairy F., Carr A., Enoch T. rqh1+, a fission yeast gene related to the Bloom's and Werner's syndrome genes, is required for reversible S phase arrest. EMBO J. 1997;16:2682–2692. doi: 10.1093/emboj/16.10.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Franchitto A., Pirzio L., Prosperi E., Sapora O., Bignami M., Pichierri P. Replication fork stalling in WRN-deficient cells is overcome by prompt activation of a MUS81-dependent pathway. J. Cell Biol. 2008;183:241–252. doi: 10.1083/jcb.200803173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pichierri P., Franchitto A., Mosesso P., Palitti F. Werner's syndrome protein is required for correct recovery after replication arrest and DNA damage induced in S-phase of cell cycle. Mol. Biol. Cell. 2001;12:2412–2421. doi: 10.1091/mbc.12.8.2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prince P., Emond M., Monnat R.J. Loss of Werner syndrome protein function promotes aberrant mitotic recombination. Genes Dev. 2001;15:933–938. doi: 10.1101/gad.877001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saintigny Y., Makienko K., Swanson C., Emond M., Monnat R.J. Homologous recombination resolution defect in werner syndrome. Mol. Cell. Biol. 2002;22:6971–6978. doi: 10.1128/MCB.22.20.6971-6978.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Swanson C., Saintigny Y., Emond M.J., Monnat R.J., Jr. The Werner syndrome protein has separable recombination and survival functions. DNA Repair (Amst.) 2004;3:475–482. doi: 10.1016/j.dnarep.2004.01.002. [DOI] [PubMed] [Google Scholar]
- 22.Kamath-Loeb A.S., Lan L., Nakajima S., Yasui A., Loeb L.A. Werner syndrome protein interacts functionally with translesion DNA polymerases. Proc. Natl. Acad. Sci. U.S.A. 2007;104:10394–10399. doi: 10.1073/pnas.0702513104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dong Y., Seki M., Yoshimura A., Inoue E., Furukawa S., Tada S., Enomoto T. WRN functions in a RAD18-dependent damage avoidance pathway. Biol. Pharm. Bull. 2007;30:1080–1083. doi: 10.1248/bpb.30.1080. [DOI] [PubMed] [Google Scholar]
- 24.Sidorova J., Nianzhen L., Folch A., Monnat R.J., Jr. The RecQ helicase WRN is required for normal replication fork progression after DNA damage or replication fork arrest. Cell Cycle. 2008;7:796–807. doi: 10.4161/cc.7.6.5566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jansen J., Tsaalbi-Shtylik A., Hendriks G., Gali H., Hendel A., Johansson F., Erixon K., Livneh Z., Mullenders L., Haracska L., de Wind N. Separate domains of Rev1 mediate two modes of DNA damage bypass in mammalian cells. Mol. Cell. Biol. 2009;29:3113–3123. doi: 10.1128/MCB.00071-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simpson L.J., Sale J.E. Rev1 is essential for DNA damage tolerance and non-templated immunoglobulin gene mutation in a vertebrate cell line. EMBO J. 2003;22:1654–1664. doi: 10.1093/emboj/cdg161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arakawa H., Moldovan G.L., Saribasak H., Saribasak N.N., Jentsch S., Buerstedde J.M. A role for PCNA ubiquitination in immunoglobulin hypermutation. PLoS Biol. 2006;4:e366. doi: 10.1371/journal.pbio.0040366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamashita Y.M., Okada T., Matsusaka T., Sonoda E., Zhao G.Y., Araki K., Tateishi S., Yamaizumi M., Takeda S. RAD18 and RAD54 cooperatively contribute to maintenance of genomic stability in vertebrate cells. EMBO J. 2002;21:5558–5566. doi: 10.1093/emboj/cdf534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arakawa H., Lodygin D., Buerstedde J.M. Mutant loxP vectors for selectable marker recycle and conditional knock-outs. BMC Biotechnol. 2001;1:7. doi: 10.1186/1472-6750-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Franklin R., Sale J.E. 2D cell cycle analysis. Subcell. Biochem. 2006;40:405–408. doi: 10.1007/978-1-4020-4896-8_35. [DOI] [PubMed] [Google Scholar]
- 31.Simpson L.J., Sale J.E. Colony survival assay. Subcell. Biochem. 2006;40:387–391. doi: 10.1007/978-1-4020-4896-8_31. [DOI] [PubMed] [Google Scholar]
- 32.Imamura O., Fujita K., Itoh C., Takeda S., Furuichi Y., Matsumoto T. Werner and Bloom helicases are involved in DNA repair in a complementary fashion. Oncogene. 2002;21:954–963. doi: 10.1038/sj.onc.1205143. [DOI] [PubMed] [Google Scholar]
- 33.Takeuchi F., Hanaoka F., Goto M., Yamada M., Miyamoto T. Prolongation of S phase and whole cell cycle in Werner's syndrome fibroblasts. Exp. Gerontol. 1982;17:473–480. doi: 10.1016/s0531-5565(82)80009-0. [DOI] [PubMed] [Google Scholar]
- 34.Sidorova J.M. Roles of the Werner syndrome RecQ helicase in DNA replication. DNA Repair (Amst.) 2008;7:1776–1786. doi: 10.1016/j.dnarep.2008.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fujiwara Y., Higashikawa T., Tatsumi M. A retarded rate of DNA replication and normal level of DNA repair in Werner's syndrome fibroblasts in culture. J. Cell. Physiol. 1977;92:365–374. doi: 10.1002/jcp.1040920305. [DOI] [PubMed] [Google Scholar]
- 36.Poot M., Hoehn H., Runger T.M., Martin G.M. Impaired S-phase transit of Werner syndrome cells expressed in lymphoblastoid cell lines. Exp. Cell Res. 1992;202:267–273. doi: 10.1016/0014-4827(92)90074-i. [DOI] [PubMed] [Google Scholar]
- 37.Poot M., Gollahon K.A., Emond M.J., Silber J.R., Rabinovitch P.S. Werner syndrome diploid fibroblasts are sensitive to 4-nitroquinoline-N-oxide and 8-methoxypsoralen: implications for the disease phenotype. FASEB J. 2002;16:757–758. doi: 10.1096/fj.01-0906fje. [DOI] [PubMed] [Google Scholar]
- 38.Prince P.R., Ogburn C.E., Moser M.J., Emond M.J., Martin G.M., Monnat R.J., Jr. Cell fusion corrects the 4-nitroquinoline 1-oxide sensitivity of Werner syndrome fibroblast cell lines. Hum. Genet. 1999;105:132–138. doi: 10.1007/s004399900078. [DOI] [PubMed] [Google Scholar]
- 39.Ogburn C.E., Oshima J., Poot M., Chen R., Hunt K.E., Gollahon K.A., Rabinovitch P.S., Martin G.M. An apoptosis-inducing genotoxin differentiates heterozygotic carriers for Werner helicase mutations from wild-type and homozygous mutants. Hum. Genet. 1997;101:121–125. doi: 10.1007/s004390050599. [DOI] [PubMed] [Google Scholar]
- 40.Poot M., Yom J.S., Whang S.H., Kato J.T., Gollahon K.A., Rabinovitch P.S. Werner syndrome cells are sensitive to DNA cross-linking drugs. FASEB J. 2001;15:1224–1226. doi: 10.1096/fj.00-0611fje. [DOI] [PubMed] [Google Scholar]
- 41.Dhillon K.K., Sidorova J., Saintigny Y., Poot M., Gollahon K., Rabinovitch P.S., Monnat R.J., Jr. Functional role of the Werner syndrome RecQ helicase in human fibroblasts. Aging Cell. 2007;6:53–61. doi: 10.1111/j.1474-9726.2006.00260.x. [DOI] [PubMed] [Google Scholar]
- 42.Harrigan J.A., Wilson D.M., 3rd, Prasad R., Opresko P.L., Beck G., May A., Wilson S.H., Bohr V.A. The Werner syndrome protein operates in base excision repair and cooperates with DNA polymerase β. Nucleic Acids Res. 2006;34:745–754. doi: 10.1093/nar/gkj475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tateishi S., Niwa H., Miyazaki J., Fujimoto S., Inoue H., Yamaizumi M. Enhanced genomic instability and defective postreplication repair in RAD18 knockout mouse embryonic stem cells. Mol. Cell. Biol. 2003;23:474–481. doi: 10.1128/MCB.23.2.474-481.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Szüts D., Simpson L.J., Kabani S., Yamazoe M., Sale J.E. Role for RAD18 in homologous recombination in DT40 cells. Mol. Cell. Biol. 2006;26:8032–8041. doi: 10.1128/MCB.01291-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gebhart E., Bauer R., Raub U., Schinzel M., Ruprecht K.W., Jonas J.B. Spontaneous and induced chromosomal instability in Werner syndrome. Hum. Genet. 1988;80:135–139. doi: 10.1007/BF00702855. [DOI] [PubMed] [Google Scholar]
- 46.Rupp W.D., Howard-Flanders P. Discontinuities in the DNA synthesized in an excision-defective strain of Escherichia coli following ultraviolet irradiation. J. Mol. Biol. 1968;31:291–304. doi: 10.1016/0022-2836(68)90445-2. [DOI] [PubMed] [Google Scholar]
- 47.Tateishi S., Sakuraba Y., Masuyama S., Inoue H., Yamaizumi M. Dysfunction of human Rad18 results in defective postreplication repair and hypersensitivity to multiple mutagens. Proc. Natl. Acad. Sci. U.S.A. 2000;97:7927–7932. doi: 10.1073/pnas.97.14.7927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guo C., Fischhaber P.L., Luk-Paszyc M.J., Masuda Y., Zhou J., Kamiya K., Kisker C., Friedberg E.C. Mouse Rev1 protein interacts with multiple DNA polymerases involved in translesion DNA synthesis. EMBO J. 2003;22:6621–6630. doi: 10.1093/emboj/cdg626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ross A.L., Simpson L.J., Sale J.E. Vertebrate DNA damage tolerance requires the C-terminus but not BRCT or transferase domains of REV1. Nucleic Acids Res. 2005;33:1280–1289. doi: 10.1093/nar/gki279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Szüts D., Marcus A.P., Himoto M., Iwai S., Sale J.E. REV1 restrains DNA polymerase ζ to ensure frame fidelity during translesion synthesis of UV photoproducts in vivo. Nucleic Acids Res. 2008;36:6767–6780. doi: 10.1093/nar/gkn651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jackson D.A., Pombo A. Replicon clusters are stable units of chromosome structure: evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J. Cell Biol. 1998;140:1285–1295. doi: 10.1083/jcb.140.6.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]