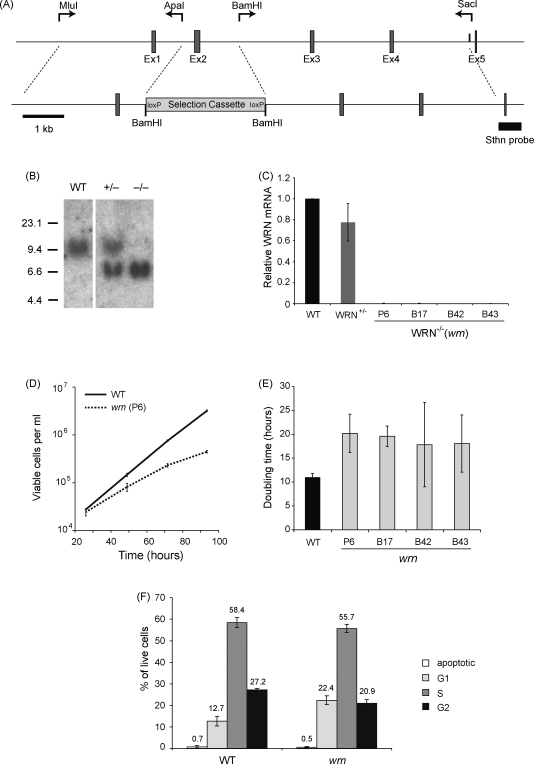
Fig. 1.
Generation of a WRN null DT40 line. (A) Schematic of the WRN gene targeting strategy. The binding sites of the primers used to generate the 5′ and 3′ arms of the targeting construct are shown above the locus map, along with the restriction sites used in their cloning. A selection cassette replaces exon 2 of the WRN transcript and which, following targeted integration, is predicted to lead to an out of frame splice event between exons 1 and 3 resulting in a stop codon 16 amino acids into exon 3. The Southern probe spanning exon 5 is indicated. (B) BamHI digested genomic DNA from wild type (WT), WRN+/−and WRN−/−(wrn) cells probed as indicated above. All lanes are from the same blot, but intervening lanes have been cut out between WT and +/−. (C) qPCR for exons 19–23 confirming loss of WRN transcript downstream of the disruption in four clones. (D) Growth of wild type (WT) and wrn clone P6. The graph represents three independent experiments with the error bars indicating the standard deviation. (E) Comparison of the doubling time in four independent clones of wrn DT40. (F) Cell cycle distribution of cells in asynchronous cultures of wild type and wrn lines. The mean percentage of 3 independent determinations is shown and the error bars represent one standard deviation.