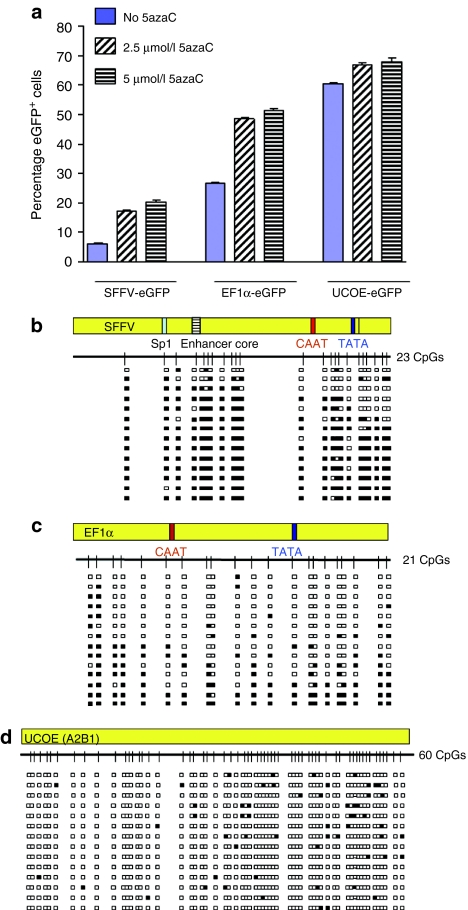
Figure 2.
A2UCOE is not subject to DNA methylation in P19 cells. (a) Abrogation of SFFV-eGFP and EF1α-eGFP silencing in P19 cells by addition of the DNA methylation inhibitor 5-azacytidine (5azaC). 5azaC was added to cultures 2.5 and 5 µmol/l on day 8 after transduction with eGFP expression assessed 5 days later. The mean and standard deviation of triplicate experiments for each vector are shown. Solid bars: no 5azaC. Hatched bars: 2.5 µmol/l 5azaC. Horizontal cross line bars: 5 µmol/l 5azaC. Note: percentage of eGFP+ cells transduced with the SFFV and EF1α vectors remained at a high level when treated with 5azaC compared to untreated cells; 5azaC treatment had a much smaller effect on eGFP expression from the A2UCOE. The P values of 5azaC treated to untreated cells: SFFV-eGFP 2.5 µmol/l: 1.9992 × 10−5, 5 µmol/l: 7.65488 × 10−6; EF1α-eGFP 2.5 µmol/l: 9.95594 × 10−7, 5 µmol/l: 5.95237 × 10−7; A2UCOE-eGFP 2.5 µmol/l: 0.000299, 5 µmol/l: 0.000143967. (b–d) DNA methylation analysis of SFFV (b), EF1α (c), and A2UCOE (d) proviral elements. Genomic DNA from cells transduced with lentiviruses at day 17 after transduction was isolated and subjected to methylation analysis by bisulfite conversion and sequencing. Methylation status of randomly selected PCR clones generated after conversion is shown. White boxes: unmethylated CpG sites. Black boxes: methylated CpG sites. Sp1: Sp1 transcription factor binding site. Enhancer core: SFFV enhancer core element; CAAT, TATA: CAAT and TATA box elements. Note: only scattered methylated CpG sites are found within the A2UCOE sequence compared to extensive methylation of CpG dinucleotides in EF1α and SFFV.