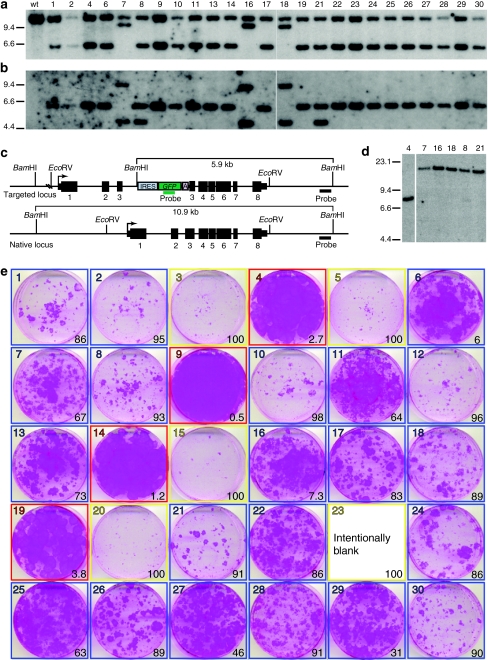
Figure 4.
Clonal analysis of KRT14-targeted normal human keratinocytes. Normal human keratinocytes were transduced with AAVK14e3IFsA and GFP positive cells sorted for clonal analysis. Thirty clones were examined for evidence of targeted insertions at the KRT14 locus, and for the growth phenotype of colonies on irradiated NIH-3T3-J2 feeders. Four of these clones were subsequently used to construct skin equivalents that were transplanted to nude mice for further analysis. (a) Southern blot analysis of BamHI digested genomic DNA from 25 clones that grew sufficiently for DNA to be collected. The blot is probed with a DNA fragment from the KRT14 locus (outside of vector homology). (b) The same blot as in a, with KRT14 probe removed and probed with a GFP gene fragment. (c) Diagram of the KRT14-targeted and native locus. Restriction enzyme sites and relevant fragment sizes are indicated as well as the position of both the KRT14 probe and the GFP probe used for Southern in a and b. Transcription start sites are indicated by bent arrows. Exons are depicted as black boxes and numbered below with introns shown as a connecting line. 5′ and 3′ untranslated regions are shown as narrow boxes. (d) Genomic DNA from clones 7, 16, 18, 8, and 21 was digested with EcoRV and probed with a GFP gene fragment to determine if multiple vector copies present in these clones were composed as tandem integrants at the KRT14 locus, or at multiple different integration sites. Genomic DNA from clone 4 is used as a control. (e) Colony forming efficiency (CFE) assay used to define which targeted clones are paraclones (100% abortive colonies, outlined in yellow), meroclones (6–99% abortive colonies, outlined in blue), or holoclones (<5% abortive colonies, outlined in red). The photographs are of 6-cm diameter culture dishes containing formaldehyde-fixed colonies stained with rhodamine B. Numbers in the lower right of each box indicate the percentage of abortive colonies on the dish as determined by colony diameter size of <4 mm after 12 days of growth. Colonies from clone 23 were pooled with cells from the other dishes to generate enough DNA for Southern analysis so no picture was obtained from this CFE assay for this clone.