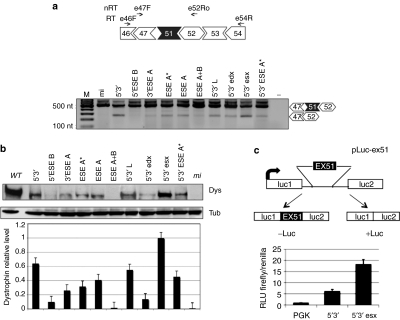
Figure 3.
Exon 51 skipping and dystrophin rescue in DMD Δ48–50 primary myoblasts and selection of the best performing construct with the Luciferase-based splicing-reporter assay. (a) Top: schematic representations of the Δ48–50 dystrophin complementary DNA between exons 46 and 54 together with the positions of reverse transcriptase-PCR oligos (E46F and E54R—RT), and nested PCR oligos (E47F and E52Ro—nRT). Bottom: nested RT-PCR on RNA extracted from Δ48–50 cells infected with the antisense-expressing lentiviruses. Unskipped and skipped products are indicated on the right. Molecular sizes are indicated on the left. (b) Representative western blot of proteins (50 µg) extracted from infected Δ48–50 cells, probed with anti-dystrophin (Dys) and anti-tubulin (Tub) antibodies (WT: 2 µg of proteins from wild-type skeletal muscle cells; mi: 50 µg of proteins from mi cells). The histogram at the bottom indicates dystrophin levels normalized on tubulin signals. Values are obtained from three independent experiments; error bars: means ± SD. (c) Top: schematic representation of the pLuc-ex51construct. Exon 51 and part of its flanking introns were cloned between the Firefly luciferase exons. From this construct, luciferase expression is obtained only upon exon 51 skipping. Bottom: histogram showing the relative luminescence units (RLU) from C27 myoblast cells transfected with pLuc-ex51 and pTK (Renilla luciferase expressing plasmid), together with either 5′–3′ or 5′–3′esx constructs. An empty lentiviral vector is used as control (PGK). DMD, Duchenne muscular dystrophy; mi, mock infected; −: negative control without RNA.