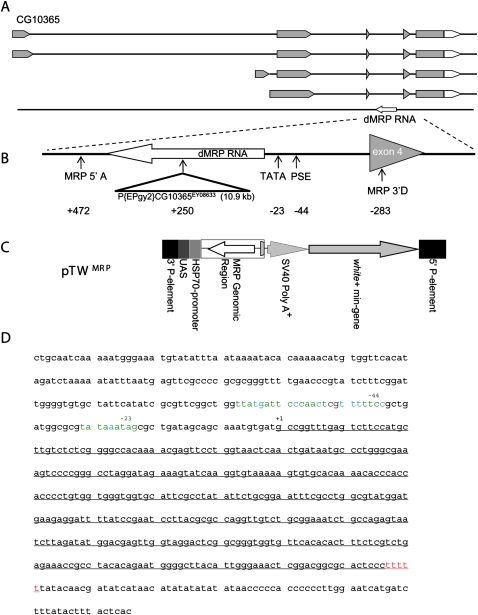
FIGURE 1.
The Drosophila (dMRP) RNA gene. (A) Piccinelli et al. (2005) predicted a potential dMRP RNA encoded by a gene located in the third intron of CG10365, transcribed from the opposite strand. (Gray) Translated sequences. (B) Expanded view of the dMRP gene. P{EPgy2} CG10365EY08633 disrupts dMRP in the dMRPEY08633 mutant strain. Positions of primers used to amplify the genomic region used to make the pTW -MRP strain (MRP 5′A and MRP 5′D) are indicated. (C) Structure of pTW MRP used to make the pTW -MRP strain. Regions inserted into chromosome 2 by untargeted integration via 5′ and 3′ P-element sequences are shown. dMRP genomic sequences were inserted in the opposite orientation relative to the UAS/Hsp70 transcriptional regulatory sequences so that dMRP RNA was expressed from its own regulatory sequences. The white (+) mini-gene encodes a red eye-color gene for selecting transgenic individuals. (D) dMRP sequence with potential Pol III regulatory elements. The sequence of the entire intron containing dMRP is shown. Transcribed sequences are underlined. Positions of PSE (−44 nt) and TATA-like element (−23 nt) upstream of the transcription start site (+1) are indicated above the sequence. (Green nucleotides in promoter elements) Identical nucleotides in all reference sequences (Hernandez et al. 2007), (blue nucleotides) the same as at least one of the reference sequences, (red nucleotides) consensus Pol III terminator (Hernandez et al. 2007).