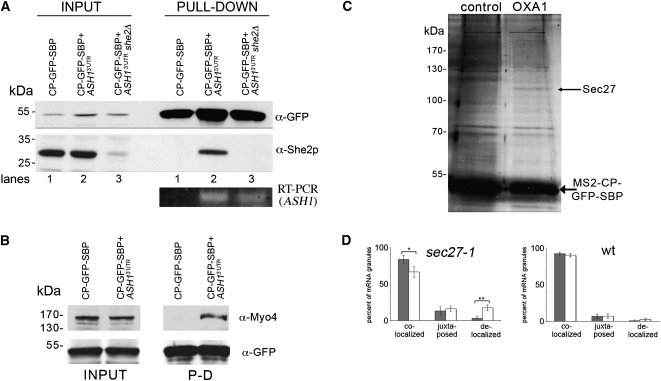
FIGURE 4.
Identification of RNA-binding proteins using RaPID. (A) Endogenous She2 interacts with the 3′UTR of ASH1 mRNA. Wild-type and she2Δ yeast expressing pMS2-CP-GFP-SBP and MS2L-tagged ASH13′UTR mRNA, or wild-type yeast expressing pMS2-CP-GFP-SBP alone were grown in 400-mL cultures, treated, lysed, and 25 mg of the total protein extract from each sample was subjected to RaPID. For Western analysis, 85% of the eluates (PULL-DOWN) and 40-μg samples of the total protein (INPUT) were resolved by SDS-PAGE and analyzed using anti-She2 and anti-GFP antibodies. Note that the faint band detected with anti-She2 antibodies in input lane 3 is probably due to recognition of a nonspecific protein. For RT–PCR analysis, RNA was isolated from the remaining 15% of the eluate and analyzed by RT–PCR with a primer pair that recognizes the ASH13′UTR. (B) Endogenous Myo4 is identified in the precipitated ASH1 mRNP complex. Yeast expressing pMS2-CP-GFP-SBP and MS2L-tagged ASH13′UTR mRNA or cells expressing pMS2-CP-GFP-SBP alone were grown in 400-mL cultures, treated, lysed, and 30 mg of the total protein extract from each sample was subjected to the RaPID procedure. The eluates (PULL-DOWN) and 40-μg samples of the total protein (INPUT) were resolved by SDS-PAGE and analyzed with anti-Myo4 and anti-GFP antibodies. (C) Identification of Sec27 as a candidate OXA1 mRNA interacting protein. Wild-type yeast expressing RFP-OXA1-MS2L (OXA1) or RFP-MS2L (control) were grown in cultures of 800 mL, treated, lysed and aliquots of 50 mg of total protein extract were processed using RaPID. Following the reversal of cross-linking, the eluate was resolved by SDS-PAGE using a 20 x15 cm 9% polyacrylamide gel, silver stained, and select bands were analyzed by mass spectrometry. The thin arrow marks Sec27, while the thick arrow marks the precipitated MS2-CP-GFP-SBP protein. (D) Inactivation of Sec27 increases the proportion of delocalized OXA1 mRNA granules. Wild-type or sec27-1 yeast strains expressing RFP-OXA1-MS2L mRNA and MS2-CP-GFP(x3) were grown to mid-log phase (O.D.600 ≅ 1) at 26°C. Cells were shifted for 1 h to medium lacking methionine at the permissive temperature (26°C) and then either shifted to the restrictive temperature (37°C) for 1 h or maintained at 26°C. The cells were fixed and the localization of OXA1 mRNA granules relative to mitochondrial structures (as labeled with Oxa1-RFP) was analyzed using confocal microscopy. The gray-filled and white (unfilled) columns of the histogram indicate the distribution of granules in cells incubated at 26oC and 37oC, respectively. Statistics show the average percentage (±SE) of colocalized, juxtaposed (within ≤0.2 μm), or delocalized granules relative to the closest mitochondrial structure. (*) P = 0.035; (**) P = 0.006.