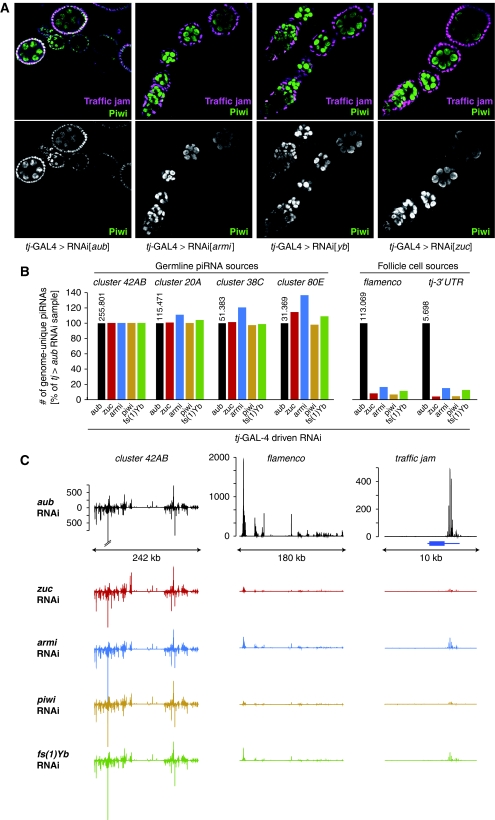
Figure 7.
piRNA biogenesis defects in somatic cells lacking Armi, Piwi, Zuc and Yb. (A) Shown are ovarioles stained for Piwi (green) and Traffic jam (magenta) from flies expressing the indicated dsRNAs with tj-GAL4. Note the lack of nuclear Piwi in follicle cells that are marked by Tj (aub knockdown serves as negative control). (B) Shown are relative levels of genome-unique Piwi–piRNAs mapping to the indicated piRNA clusters in ovaries of the indicated genotypes. Libraries were normalized through the genome-unique mappers to 42AB, a germline-specific piRNA cluster. The tj-GAL4>aub-RNAi sample serves as control. The respective raw read numbers from the tj>aub_RNAi library are indicated above the black bars. (C) Detailed piRNA profiles (only genome-unique piRNAs are shown) across the piRNA clusters 42AB and flamenco as well as the major genic piRNA precursor tj. The colour code is as in (B) and the annotated tj gene is indicated in blue. The y axis indicates the number of sequences per 200 nt window normalized to 1 million sequenced reads and graphs within a vertical row are shown at the same scale.