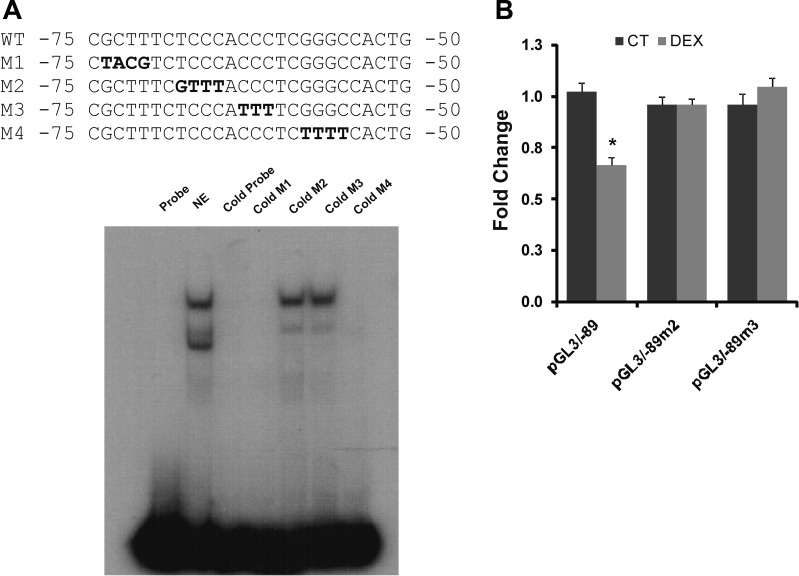
Fig. 6.
Identification of the DNA sequences on the proximal promoter region of the human NHE8 gene involved in the dexamethasone regulation. A: identification of the DNA sequences involved in dexamethasone regulation. DNA oligo 3 was end-labeled with [32P]ATP and used as a probe in GMSAs. Nuclear protein was isolated from Caco-2 cells, and GMSAs were performed as described in materials and methods. Mutant oligos (oligos M1, M2, M3, and M4) were used to identify the precise DNA sequences of the DNA-protein interaction. Bold letters indicate the mutation site at the promoter region. GMSA image indicates that cold probe and mutant probes (M1, M4) compete with the protein binding on the labeled oligo 3 probe, but mutant oligos (M2 and M3) could not compete with the protein binding on the labeled oligo 3 probe. B: functional characterization of DNA-protein interacting sequences at NHE8 promoter region. Wild-type (WT) promoter construct (pGL3b/−89) or mutant promoter constructs (pGL3b/−89M2 and pGL3b/−89M3) were cotransfected with pRL-CMV in Caco-2 cells. Promoter reporter assay was performed 18 h after dexamethasone treatment. Promoter activity was calculated as relative activity of firefly luciferase activity driven by NHE8 promoter to Renilla luciferase activity driven by CMV promoter. Fold changes were used to compare the promoter activity in the dexamethasone group with that of the control group. Results are means ± SE from 4 separate experiments. *P ≤ 0.05 for control treatment (CT) vs. dexamethasone treatment.