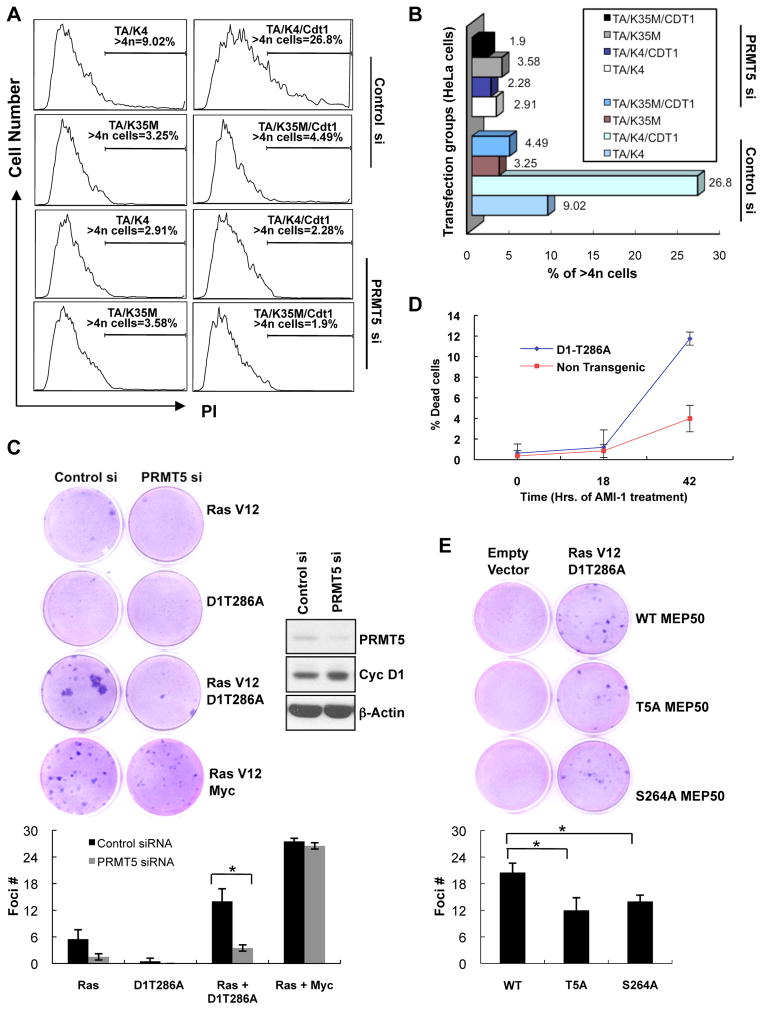
Figure 6. PRMT5 knockdown inhibits cyclin D1T286A-dependent DNA re-replication, cell transformation and increases death of tumor cells.
(A) Following transfection of HeLa cells with the indicated expression plasmids, TA (D1T286A), K4 (CDK4), K35M (CDK4(K35M)), CDT1, cell cycle profile was assessed by FACS after PI staining. (B) Graphical representation of the flow cytometry data from (A). The bars show the percentage of cells showing >4N DNA content. (C) Foci formation assay was performed by concurrent knockdown of murine PRMT5 and overexpression of the indicated proteins in NIH3T3 cells. Cells were grown for 14 days and stained with Giemsa to visualize foci. Quantification of data has been shown. Error bars represent ±SD and * indicates p-value < 0.05. (D) Single cell suspensions of splenocytes prepared from non-transgenic or malignant Eμ-D1T286A transgenic tumor burdened spleens were treated with 200μM arginine methyltransferase inhibitor AMI-1. The cells were analyzed for the inhibitor sensitivity by measuring the % cell death via AnnexinV –PI staining/flow cytometry. Values represent the mean of three independent experiments with error bars indicating ±SD. (E) Foci formation assay was performed by transfection of the indicated MEP50 constructs along with empty vector control or RasV12 plus cyclin D1T286A in NIH 3T3 cells. Cells were grown for 14 days and foci were analyzed as in (C). See also Figure S5.