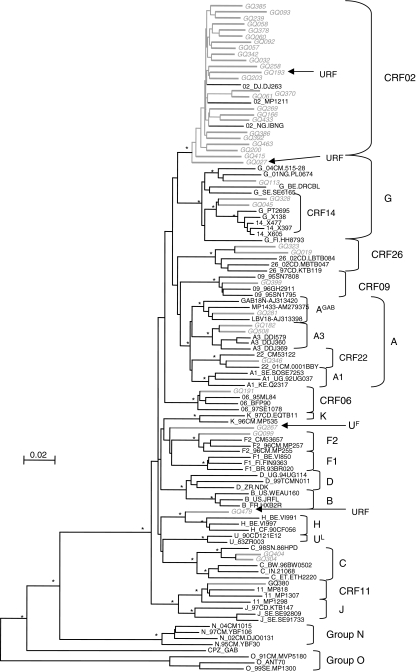
FIG. 2.
Phylogenetic relationships of the pol (protease + RT) sequences from the 41 amplified HIV-1 group M samples from the study population in Equatorial Guinea. The phylogenetic tree analysis was done using the neighbor joining (NJ) method with 100 bootstrap resamplings on 1529 unambiguously aligned nucleotides from the gap-stripped alignment. Sequences from Equatorial Guinea are highlighted in gray and bootstrap values above 90% are indicated with an asterisk (*). The strains identified as unique recombinants after Simplot and bootscan analysis were noted as “URF” and indicated with an arrow.