Abstract
The study of temperature sensitive (Ts) mutant phenotypes is fundamental to gene identification and for dissecting essential gene function. In this chapter we describe two “shuffling” methods for producing Ts mutants using a combination of PCR, in vivo recombination, and transformation of diploid strains heterozygous for a knockout of the desired mutation. The main difference between the two methods is the type of strain produced. In the “plasmid” version, the product is a knockout mutant carrying a centromeric plasmid carrying the Ts mutant. In the “chromosomal” version, The Ts allele is integrated directly into the endogenous locus, albeit not in an entirely native configuration. Both variations have the ir strengths and weaknesses, which are discussed here.
Introduction
The study of temperature sensitive (Ts) mutant phenotypes has proven to be a fundamental approach both for the identification of gene sets essential for various aspects of biology and for obtaining a detailed understanding of essential gene function. While the observation that temperature sensitive mutations represent a general class of mutation was recognized in the ‘50s (Horowitz, 1950), the first targeted screen, isolation, and analysis of Ts mutants (382 mutations located in 37 genes scattered widely over the bacteriophage T4 genome) was by Edgar and Lielausis in 1963 (Edgar and Lielausis, 1964). Lee Hartwell, in 1967, reported the isolation of 400 Ts mutations in S. cerevisiae, which caused defects in essential processes including cell division, and protein, RNA, and DNA synthesis (Hartwell, 1967). Over the past 40 years, the isolation and analysis of Ts mutations in essential genes has been a linchpin technology for investigating the genetics and molecular biology of essential processes in all experimental organisms.
Ts mutations are typically missense mutations, which retain the function of a specific essential gene at standard (permissive) low temperature, lack that function at a defined high (non-permissive) temperature, and exhibit partial (hypomorphic) function at an intermediate (semi-permissive) temperature. Such mutants make possible the analysis of physiologic changes that follow controlled inactivation of a gene or gene product by shifting cells to a non-permissive temperature, offering a powerful approach to the analysis of gene function.
Essential genes, by definition, encode critical cellular functions that are not buffered by redundant functions or pathways (Hartman et al., 2001). Essential genes have been shown to be highly dense hubs within genetic interaction networks and are involved in all aspects of basic cellular function (Jeong et al., 2001). Furthermore, essential genes tend to be more highly conserved in evolution; 38% of essential yeast proteins have easily identifiable counterparts in humans, versus 20% for nonessential genes (Hughes, 2002).
Despite their importance, the functions of many essential yeast proteins have not been studied. In part, this is due to the absence of essential gene representation in the genome wide haploid mutant collections, which cover all of the ~5000 non-essential yeast genes. Thus, no comparable systematic haploid mutant collection currently exists for the ~1000 essential genes in S. cerevisiae. The frequency of sites mutable to a reduced or conditional function is highly gene-specific; for example, for highly conserved proteins, random single missense mutation would be expected, for the vast majority of positions within the protein, to cause complete loss of function when mutated. Therefore, genetic screens using a random mutagenesis approach rarely reach saturation because “mutability” varies widely among genes.
Here we report detailed protocols for two methodologies that allow the systematic isolation of Ts alleles in essential genes of interest. The first method is plasmid-based, and the second is genome integration based, and each has its specific advantages depending on the application. Both methods exploit features of the “haploid convertible” heterozygous diploid collection, which allows introduction of the library of mutagenized essential gene copies into the heterozygous diploid and subsequent direct selection of haploids which are deleted for the target essential gene and that carry individual members of the mutagenized essential gene library, using the “diploid shuffle” technique (see below). Several other useful corollary methods for transferring extant Ts alleles or specific gene constructs (eg, fusion proteins) are also presented. Finally, we note that our laboratories are in the process of generating a complete set of Ts alleles for each of the essential genes in S. cerevisiae, which will be distributed as a resource to the scientific community when completed (see Ben Aroya etal 2008 for details). For specific essential genes under study in individual laboratories, however, it may be useful, using the methods described here, to generate an additional series of independent Ts alleles for detailed functional analysis. Furthermore, mutagenized libraries of specific essential or non-essential genes can be screened for conditional viability under a variety of conditions that are normally sub-lethal in the wildtype strain (eg, sublethal doses of drugs) using the methods described here.
Part I. Diploid shuffle – plasmid method
General description of the diploid shuffle – plasmid method
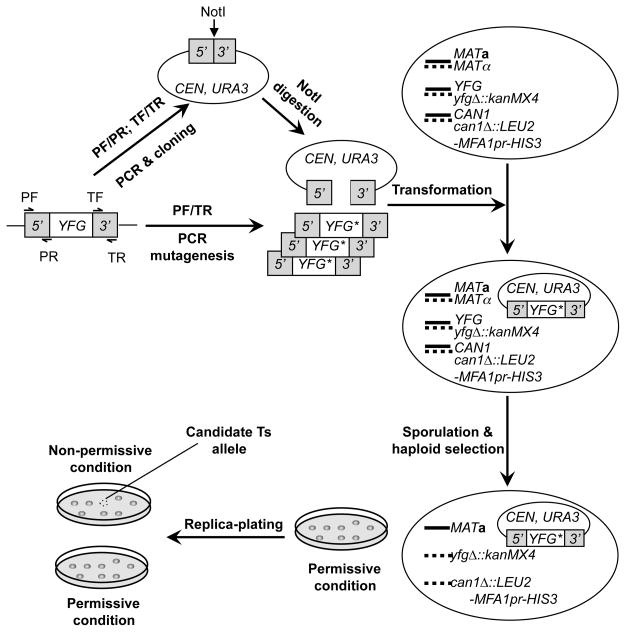
Much like traditional plasmid-shuffling methods, mutants generated by this version of the diploid shuffle are plasmid-borne alleles that can be very easily transferred to and tested in different strain backgrounds. The experimental procedure is outlined in Fig. 1. First, the endogenous promoter (including the 5′ untranslated region, 5′UTR, ~500bp) and the terminator (3′UTR, ~500bp) of a gene of interest (or your favorite gene, YFG) are PCR amplified and cloned in tandem onto a centromere-based yeast-E. coli shuttle vector, which contains URA3 as the selectable marker in yeast cells, such as pRS416. The resultant promoter/terminator clone is subsequently linearized with an endonuclease, typically NotI, which cuts at a site pre-engineered between the promoter and terminator. Simultaneously, the sequence of YFG, including the whole open reading frame (ORF), complete with the promoter and terminator regions, is randomly mutagenized with error prone PCR. The linearized promoter/terminator plasmid and PCR products are then combined and co-transformed into a haploid-convertible heterozygous diploid deletion mutant (YFG/yfgΔ::kanMX4) in the same gene being mutagenized. The mutagenized PCR product is thereby cloned into the URA3 plasmid via recombination mediated by the terminal homologous DNA sequences of both the PCR products and the linearized vector. The Ura+ transformants are subsequently cultured in a sporulation medium and converted into haploid cells by growing on a medium that allow growth of only haploid MATa G418R Ura+ cells. In these cells, the chromosomal wild-type copy of YFG is deleted, allowing direct observation of any phenotypes of the plasmid-borne alleles. To screen for conditional alleles, such haploid cells are first grown under a permissive condition such as low temperature. Colonies formed are subsequently replica-plated to fresh plates at permissive and nonpermissive conditions. Conditional alleles are identified as those grow under the permissive but not the non-permissive condition and subsequently verified. This method has been used to create thermosensitive (Ts) alleles of multiple essential genes as well as a large collection of methyl methanesulfonate (MMS) hypersensitive alleles of POL30 (Huang et al., 2008; Lin et al., 2008). Here we will outline the detailed methods to generating and verifying Ts alleles of an essential gene.
Fig. 1. A schematic for creating conditional alleles using plasmid-chromosome shuffle.
The promoter (5′) and terminator (3′) of YFG are separately PCR-amplified with primer pairs PF/PR and TF/TR, respectively, and cloned together onto a centromeric (CEN) yeast-E. coli shuttling vector. Here PF stands for promoter forward, PR for promoter reverse, TF for terminator forward, and TR for terminator reverse. A NotI recognition site is engineered between the promoter and terminator. The resultant promoter/terminator clone is linearized with NotI digestion. In the meanwhile, the entire sequence of YFG gene, including the coding region and the promoter and terminator sequences, is mutagenized using error-prone PCR with the primer pair PF/TR. The mutagenesis PCR products and the linearized promoter/terminator plasmid DNA are mixed and transformed together into a haploid-convertible heterozygous diploid knockout mutant of the same gene (MATa/α YFG/yfgΔ::kanMX4 CAN1/can1Δ::LEU2-MFA1pr-HIS3). The linearized promoter/terminator clones are repaired inside yeast cells mostly via homologous recombination using the co-transformed mutagenesis products (or YFG* alleles) as the templates. Due to the extensive homology between the ends of the PCR product and the vector, >105 recombinant clones can be easily generated. This pool of recombinants are then sporulated. Haploid MATa G418R Ura+ cells are selected under a permissive condition on solid SC-Ura-Leu-His-Arg+G418+Can medium as single colonies, which are replica-plated onto two fresh plates and incubated under both permissive and non-permissive conditions. Candidate alleles will grow under the permissive condition but not the non-permissive condition.
Materials
Media
Haploid selection synthetic medium SC–Ura–Leu–His–Arg+G418+Can: dextrose, 20 g/L; yeast nitrogen base without amino acids and ammonium sulfate, 1.7 g/L; SC–Ura–Leu–His–A dropout mix, 2g/L; sodium glutamate, 1g/L; G418, 200 mg/L; L-canavanine (Sigma, Cat# C1625), 60 mg/L; Agar, 2%. The sodium glutamate is substituted for ammonium sulfate as the nitrogen source and makes the G418 selection more reliable on the minimal medium.
SC–Ura: dextrose, 20g/L; yeast nitrogen base without amino acids and ammonium sulfate, 1.7g/L; SC–Ura dropout mix, 2g/L; ammonium sulfate, 5g/L; Agar, 2%.
Liquid YPD: yeast extract, 10g/L; peptone, 20g/L; dextrose, 20g/L.
Solid and liquid sporulation medium: potassium acetate, 10g/L; zinc acetate 0.05g/L, with or without 2% agar respectively.
Solid Luria Broth (LB) plus carbenicillin: yeast extract, 10g/L; Tryptone, 5g/L; sodium chrolide, 10g/L; carbenicillin, 50mg/L; Agar, 2%.
Liquid LB plus ampicillin: yeast extract, 10g/L; Tryptone, 5g/L; sodium chrolide, 10g/L; ampicillin, 50mg/L.
Yeast Strains
The haploid-convertible heterozygous diploid knockout mutants (MAT aα/ura3Δ0/ura3Δ0 leu2Δ0/leu2Δ0 his3Δ1/his3Δ1 lys2Δ0/LYS2 met15Δ0/MET15 can1Δ::LEU2-MFA1pr::His3/CAN1 YFG/yfgΔ::KanMX; OpenBiosystems Cat# YSC4428) (Pan et al., 2006) is used to screen for Ts alleles. Chemically competent DH5α cells prepared as described (Inoue et al., 1990) are used for cloning and plasmid recovering from yeast.
Plasmids
It is essential for this method to use a plasmid vector that contains the YFG promoter and terminator separated by a unique endonuclease (typically NotI) recognition site. Due to the limited auxotrophic markers available in the haploid-convertible heterozygous diploid knockout mutants, we normally use plasmids containing URA3 as the selectable marker such as pRS416 (Brachmann et al., 1998; Sikorski and Hieter, 1989) and YCplac33 (Gietz and Sugino, 1988). Other URA3 CEN vectors should also work.
Yeast genomic DNA
A genomic DNA sample isolated from the wild-type yeast strain BY4743 MATa/α (Brachmann et al., 1998) was used as the template for cloning the promoter and terminator of YFG and for mutagenizing its entire sequence with PCR.
Methods
Constructing the promoter/terminator clone
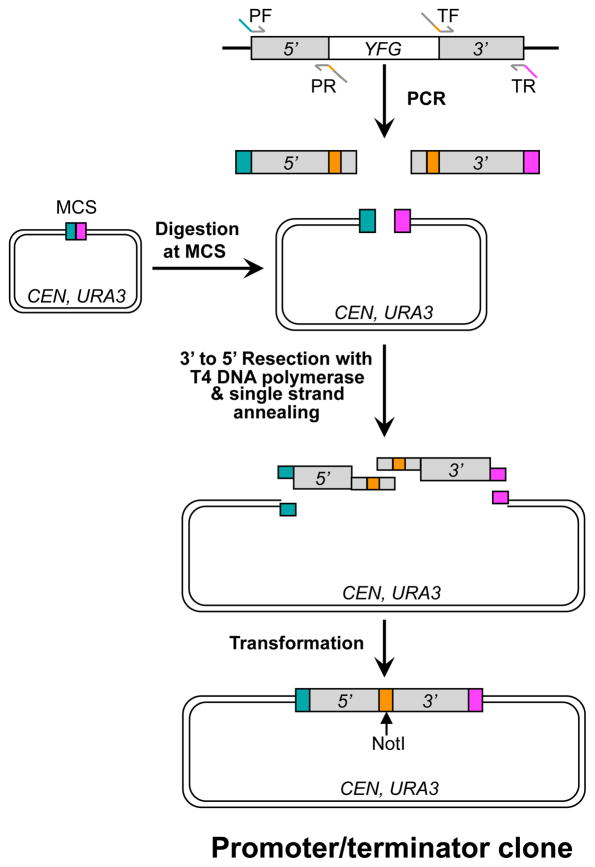
In the past, we mostly used endonuclease restriction enzyme digestion and ligation to construct the promoter/terminator clone. First, the promoter and terminator of YFG are separately PCR-amplified using primers that contain endonuclease recognition sites, for example, HindIII/NotI for the promoter and NotI/BamHI for the terminator. The PCR products are then digested with HindIII/NotI and NotI/BamHI, respectively, and ligated to pRS416 (or YCplac33) digested with HindIII/BamHI in a 3-piece ligation reaction. The ligation products are transformed into DH5α competent cells and candidate clones are selected on solid LB plus carbenicillin. More recently, we have adopted a modified version of the sequence and ligation independent cloning (SLIC) procedure (Li and Elledge, 2007) (Figure 2). This method does not require endonuclease digestion of the inserts and thus greatly simplifies primer designs and experimental procedures, especially when a large number of genes are processed simultaneously. This is also a relatively new cloning technique and is thus described below in greater detail.
Fig. 2. Constructing a promoter/terminator clone using sequence and ligation independent cloning (SLIC).
The promoter (5′) and terminator (3′) of YFG are separately PCR-amplified from yeast genomic DNA with primer pairs PF/PR and TF/TR, respectively. In the meanwhile, a centromeric yeast-E. coli shuttling vector is linearized with endonuclease digestion at the multicloning site (MCS). The PCR products and the linear vector plasmid are mixed together and processed with T4 DNA polymerase to create 5′ single-stranded overhangs. The PCR primers are designed in such a way that the PCR products and the vector can be assembled via a homology-mediated single strand annealing process. A NotI site is engineered between the cloned promoter and terminator. An aliquot of the annealing reaction is transformed into E. coli competent cells. This is a modified version of the SLIC procedure originally described by Li and Elledge (2007).
1. Design four PCR primers: promoter forward (PF), promoter reverse (PR), terminator forward (TF), and terminator reverse (TR). In addition to gene specific sequences on their 3′ termini, the PF and TR primers each contains a 30 bp sequence at the 5′ end that is either identical or complimentary to the ends of pRS416 (or YCplac33) linearized at the multicloning site (Fig. 2). The PR and TF primers are completely complementary and both contain three parts: ~20 bp promoter- or terminator-specific sequences on both ends and a NotI recognition site (8 bp) in the middle (Fig. 2).
2a. PCR-amplify the promoter and terminator according to the conditions listed in Table 1. Here genomic DNA from BY4743a/α is used as the PCR template. Platinum Pfx DNA polymerase (Invitrogen, Cat# 11708) is used due to its relative robustness and high fidelity. Others enzymes with similar features such as the Phusion (New England Biolabs, NEB, Cat# F-540) and KOD (Novagen, Cat# 71085-3) DNA polymerases can also be used.
2b. Digest ~1ug of pRS416, YCplac33, or any other URA3 plasmid with an endonuclease at the multicloning site to generate ends that are competent in cloning the PCR products via SLIC.
3. Gel-purify the PCR products and the digested vector DNA using a gel extraction kit (Qiagen, Cat# 28706 or equivalent) by following the manufacturer’s instruction. Here the PCR products of both the promoter and terminator are combined during purification. Elute each purified sample in 25μl of provided elution buffer.
4. Set up a reaction according to Table 2. Here DNA resection in the 3′ to 5′ direction (by T4 DNA polymerase, NEB, Cat# M0203) and annealing between complementary single strand overhangs occurs in the same reaction.
5. Immediately use 2μl of the above reaction to transform 20μl of chemically competent DH5α cells by following a standard protocol. This includes incubating the cell/DNA mixture sequentially on ice for 30 minutes, at 42°C for 90 seconds, back on ice for 2 minutes, and at 37°C for 30 minutes (in the presence of 200 μl of liquid LB medium).
6. Plate 100μl of the transformation mixture on solid LB plus carbenicillin to select for single colony transformants.
7. Screen for positive clones using colony-PCR with the PF and PR primers according to Table 1 but with subtle modifications. Here cells from single colonies instead of yeast genomic DNA is used to provide PCR templates.
8. Prepare plasmid DNA samples from 2–3 positive clones using a mini-spin kit (Qiagen, Cat# 27106 or equivalent) by following the manufacture’s instructions.
9. Verify each plasmid with DNA sequencing by using two primers that read towards the inserts (promoter and terminator) in both directions from the vector backbone.
Table 1.
PCR amplification of promoters and terminators.
| Component | Volume/reaction |
|---|---|
| 10 × Platinum Pfx DNA Polymerase buffer | 2.5 μl |
| dNTP (2.5 mM each) | 2 μl |
| Primer mix (5 μM each) | 2 μl |
| Yeast genomic DNA (~200 ng/μl) | 1 μl |
| ddH2O | 17.25 μl |
| Platinum Pfx DNA Polymerase (2.5 units/μl) | 0.25 μl |
| Total | 25 μl |
PCR conditions: 94°C 4 min; 30 × (94°C for 30 sec, 55°C for 30 sec, 72°C for 45 sec); 72°C for 7 min; hold at 4°C.
Table 2.
A T4 DNA polymerase resection reaction.
| Component | Volume |
|---|---|
| 10 × NEB BamHI Buffer | 2 μl |
| 10 × BSA | 2 μl |
| ddH2O | 3.7 μl |
| T4 DNA polymerase (3 units/μl) | 0.3 μl |
| Vector (100 ng/μl) | 2 μl |
| PCR product (50 ng/μl) | 10 μl |
| Total | 20 μl |
Upon set up, the reaction is incubated at 25°C for 30 minutes. After that, sit it on ice until E. coli transformation.
Mutagenizing YFG with error prone PCR
Mutagenesis of YFG using error-prone PCR is performed essentially as described previously (Leung et al, 1989). Again, a genomic DNA sample of the wild-type yeast strain BY4743a/α is used as the DNA template. The PF and TR primers described above are used to amplify the full-length gene and ~500bp flanking sequences. TaKaRa Ex Taq (Cat# RR001A) or LA Taq (Cat# RR002B), which are ~4 times more accurate than the normal Taq polymerase, are used here due to their robustness. Induction of mutation rates is achieved by adding Mn2+ in the PCR at a final concentration of 10–150 μM that are arbitrarily defined, with higher concentrations for smaller genes (genes’ sizes range from 0.5–5kb).
Set up four independent reactions for each gene according to Table 3 to reduce potential founder effects.
After PCR, pool the samples.
Examine the PCR products by agarose gel electrophoresis by using a 1–2 μl sample to ensure successful PCR amplification.
Table 3.
Error-prone PCR conditions.
| Component | Volume/reaction |
|---|---|
| 10 × Ex Taq buffer | 5 μl |
| dNTP (2.5 mM each) | 4 μl |
| Primer mixture (5 μM each) | 4 μl |
| Yeast genomic DNA (~200 ng/μl) | 2 μl |
| MnCl2 (1 to 15 mM) | 0.5 μl |
| ddH2O | 34.25 μl |
| Ex Taq DNA polymerase (5 units/ml) | 0.25 μl |
| Total | 50 μl |
PCR conditions: 94°C 4 min; 30 × (94°C for 30 sec, 55°C for 30 sec, 72°C for 1 min/kb); 72°C for 7 min; hold at 4°C. Lower MnCl2 concentrations are used for larger genes and higher concentrations for smaller genes.
Linearizing plasmid DNA of the promoter/terminator clone
Two μg of plasmid DNA of the promoter/terminator clone is digested with NotI (NEB, Cat# R0189) in NEB buffer 3 in a 20μl reaction. The reaction is incubated at 37°C for an overnight and subsequently at 65°C for 20 minutes to inactivate NotI. A small aliquot of the digestion product is examined by agarose gel electrophoresis to ensure complete digestion of the plasmid.
Combining and concentrating the PCR products and digested vector
The mutagenized PCR products (approximately 10–20 μg in 200 μl) and linearized plasmid DNA of the promoter/terminator clone (~2 μg) are next combined and concentrated by ethanol precipitation.
Transfer both the mutagenesis PCR products and NotI-digested promoter/terminator plasmid DNA into a 1.7 ml microcentrifuge tube and adjust volume to ~200 μl by adding ddH2O.
Add 5 μl of 4M ammonium acetate (pH 7.0) and 500 μl of 100% ethanol to the DNA sample and mix well by briefly vortexing. Place on ice for 10 minutes.
Precipitate DNA by spinning at >12,000rpm in a microcentrifuge for 7 minutes. There should be a tiny whitish DNA pellet at the bottom of the tube.
Carefully aspirate the liquid and wash the DNA pellet once with 300 μl of 70% ethanol.
Spin at >12,000rpm in a microcentrifuge for 3 minutes and carefully aspirate ethanol.
Dry the DNA pellet in a speed vac.
Resuspend DNA in 28 μl of sterile ddH2O.
Transforming yeast cells
The concentrated DNA sample of PCR products and the linearized vector is next transformed into the corresponding haploid-convertible heterozygous diploid yeast knockout mutant to create a mutagenized library of YFG.
Inoculate the haploid-convertible YFG/yfgΔ::kanMX4 heterozygous diploid mutant into 5 ml liquid YPD and incubate at 30°C for overnight in a roller drum.
Transfer an aliquot of the overnight culture into 50 ml YPD liquid (starting at 0.125 OD600nm/ml in a 250 ml Erlenmeyer flask) and incubate at 30°C with shaking (at 200 rpm) until a cell density of ~0.5 OD600nm/ml.
Harvest the culture in a 50 ml conical tube by spinning at 4000 rpm for 3 minutes in a bench top centrifuge and discard the medium.
Resuspend cells in 10 ml of sterile ddH2O, centrifuge as described in the previous step, and discard the supernatant.
Resuspend cells in 10 ml of 0.1 M lithium acetate (LiOAc), centrifuge, and discard the supernatant as before.
Resuspend cells in residual 0.1 M LiOAc in a total volume of 100 μl in a 1.7 ml microcentrifuge tube.
Make a transformation mixture in this order and mix well: 480 μl of 50% polyethylene glycol (PEG-3350, JTBaker, Cat# JTU221-9), 72 μl of 1M lithium acetate, 40 μl of heat-denatured herring sperm DNA (10mg/ml, Sigma, Cat# D6898) and 28 μl of DNA sample to be transformed (error-prone PCR products and linearized promoter/terminator clone).
Add the transformation mixture into the yeast competent cells prepared in step 6 and immediately mixed well by pipetting with a P1000 pipettor followed by vortexing (VMR, Vortexer 2) at top speed for 5–10 seconds.
Incubate the transformation reaction in a 30°C incubator for 30 minutes.
Add 72 μl of dimethyl sulfoxide (DMSO; Qbiogene DMSO0001, Molecular Biology Grade) to the transformation reaction and immediately mixed thoroughly by vortexing at top speed for 5–10 seconds. DMSO is intrinsically sterile and no further sterilization is needed.
Incubate the transformation reaction in a 42°C water bath for 13 minutes. A
Spin down cells at 3600 rpm in a microcentrifuge for 30 seconds to pellet cells.
Aspirate the supernatant and resuspend cells in 1 ml of sterile ddH2O.
Take 0.2 μl (1/5000) of the resuspended cells and plate on a SC–Ura plate to determine transformation yield. A successful reaction typically yields a library of 105 to 106 independent Ura+ transformants, with >90% being the recombinants between the PCR products and the linearized plasmid DNA of the promoter/terminator clone. The remainder are primarily empty vector molecules that were never cut in the first place or rejoined by nonhomologous end joining.
Sporulation
The rest of the yeast transformation reaction is either incubated in 50 ml of fresh liquid SC–Ura at 30°C for 2 days to allow propagation of the library or can be sporulated immediately to convert the transformants into a library of haploid spores that harbor mutant alleles of YFG on a plasmid (see below).
Grow the yeast transformants nonselectively in 50 ml of liquid YPD by incubating at 30°C with shaking (180–200 rpm) for 3 hours to refresh cells.
Harvest cells in a 50 ml conical tube by spinning at 4000 rpm for 3 minutes in a bench top centrifuge and discard the medium.
Resuspend cells in 40 ml of sterile ddH2O and spin at 4000 rpm for 3 minutes.
Discard the supernatant and resuspend cells in 50 ml of liquid sporulation medium.
Incubate the sporulation culture in a 250 ml flask at 25°C for 4–6 days with shaking (180–200rpm). This will typically give rise to 20–40% of sporulation efficiency when checked under a microscope.
Repeat Steps 2 and 3.
Discard the supernatant and resuspend cells in 10 ml of sterile ddH2O.
Spread aliquots (200 μl) of 10 × serial dilutions of the sporulation culture onto individual plates of the haploid selection medium SC–Ura–Leu–His–Arg+G418 +Can and incubate at 25°C for 2–3 days to determine the efficiency of producing MATa G418R Ura+ haploid cells.
Store the rest of the spores in ddH2O at 4°C for later use. The viability of spores can be maintained for a few weeks in this way.
Screening for Ts alleles
After the titer of MATa G418R Ura+ haploid cells is determined, the library is screened for potential Ts mutants. We typically screen ~4,000 clones for each gene.
Spread the spores on solid SC–Ura–Leu–His–Arg+G418 +Can, aiming for a density of ~400 MATa G418R Ura+ haploid colonies formed on each of ten plates. Store the rest of the spores at 4°C as a backup.
Incubate the plates at 25°C for 3 days to allow formation of colonies of ~2 mm in diameter.
Replica-plate the colonies from each plate to two fresh plates of the same haploid selection medium and mark orientations of the plates. It is best to pre-warm the “nonpermissive” plate to 37°C prior to replica plating.
Incubate one of the daughter plate at 25°C and the other at 37°C for 1 day.
Compare growth of each colony on both plates to assess potential Ts phenotype and select alleles that form relatively robust colonies at 25°C but ghost colonies at 37°C as candidate Ts mutants. If no Ts colony is detected, one may need to screen more clones by using the backup spores stored at 4°C, by using 38°C as the restrictive temperature, or both.
Confirming Ts mutants
Candidate mutants are picked and re-streaked onto the same haploid selection media and re-tested for the Ts phenotypes by incubating at 25°C and 37°C. The plasmids are next recovered from those confirmed to be Ts in this initial assay and re-introduced individually into the same haploid-convertible heterozygous diploid mutant to test whether the Ts phenotype is linked to the plasmids.
Grow each candidate Ts mutant in 1.5 ml of liquid SC–Ura at 25°C until saturated, typically takes 1–2 days, depending on the particular alleles.
Harvest cells of each strain in a microcentrifuge tube by spinning at 4000 rpm for 1 minute and discard the medium.
Resuspend cells in 500–1000 μl of sterile ddH2O and repeat Step 2.
Resuspend cells in 40 μl Lysis Buffer (50 mM Tris-HCl 7.5, 10 mM EDTA) containing 5 mg/ml of Zymolyase 100T (MP Biomedicals, Cat# 320931) and incubate at 37°C for 1 hour with shaking at 15-minute intervals.
Add 40 μl of 10% SDS and mix well by vortexing or pipetting.
Add 160 μl of 7.5M ammonium acetate and mix well by vortexing or pipetting.
Incubate the sample at −80°C for 15 minutes.
Centrifuge at >12,000rpm for 5 minutes at 4°C.
Carefully transfer 100 μl of clear supernatant to a new tube that contains 75 μl of isopropanol and mix well by inverting the tube several times.
Centrifuge at >12,000rpm for 7 minutes at room temperature to precipitate DNA.
Wash the DNA pellet once with 100 μl 70% ethanol.
Carefully aspirate ethanol and dry the DNA pellet in a speed vac.
Resuspend DNA pellet in 20 μl of sterile ddH2O.
Use 2 μl of the DNA sample to transform chemically competent DH5α cells as mentioned in the “constructing the promoter/terminator clone” section.
Purify plasmid DNA from a representative bacterial transformant using a kit (Qiagen, Cat# 27106 or equivalent).
Individually transform each plasmid into the haploid-convertible heterozygous diploid knockout mutant as described previously (Gietz et al., 1995) and select for Ura+ transformants.
Patch two representative Ura+ transformants for each plasmid on solid SC–Ura and incubate at 30°C for overnight.
Transfer cells to a sporulation plates and incubate at room temperature for 4–6 days.
Resuspend cells from each sporulated culture in sterile ddH2O.
Spot aliquots of 10x serial dilutions onto two SC–Ura–Leu–His–Arg+G418+Can plates.
Incubate one plate at 25°C and the other at 37°C for 2–3 days.
Compare growth of MATa G418R Ura+ cells at both temperatures. True Ts alleles will form colonies at 25°C but not at 37°C.
Part II. Diploid shuffle – chromosome method
A general description
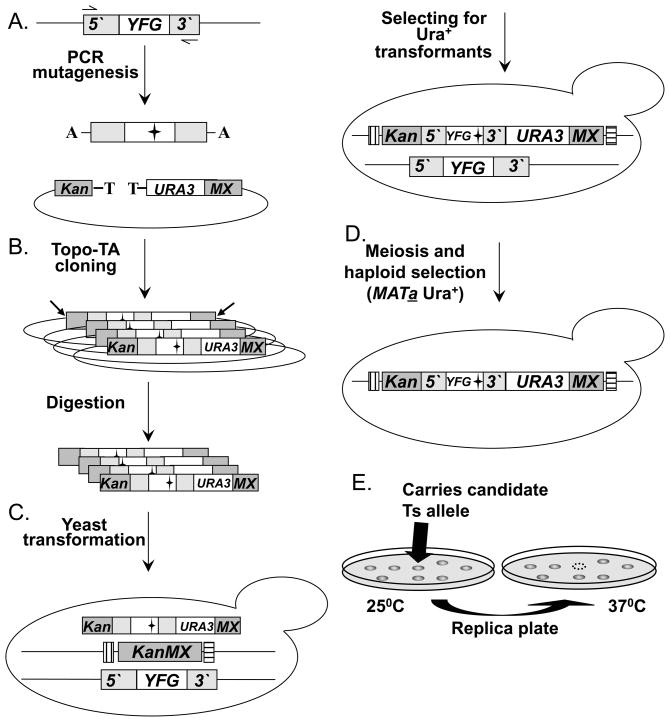
The “diploid shuffle” chromosome method has now been used to systematically screen for missense mutations that result in temperature sensitive (Ts) alleles of hundreds of essential genes, with each allele directly integrated at its endogenous chromosomal location and flanked with the “barcodes” of the corresponding yeast knockout mutant (Fig 3; (Ben-Aroya et al., 2008), and unpublished data). First, YFG, including its promoter and terminator regions, is mutagenized with error-prone PCR (Fig. 3A). The mutagenized PCR product is next cloned into SB221+Topo-TA (Fig. 3B). This plasmid contains the URA3 gene flanked by the 5′ and 3′ regions of KanMX. The Topo-TA cloning site (Invitrogen) has been inserted in between the KanMX 5′ end and the URA3 gene. This site allows direct cloning of each of the PCR products, without the need for any further modifications. The result of the cloning step is a library of mutagenized YFG, which is then transformed into E. coli, and digested to release linear fragments (following DNA purification) (Fig. 3B). The linear fragments are directly transformed into the corresponding strain from the haploid-convertible heterozygous YFG/yfgΔ::kanMX diploid YKO collection by selecting for Ura+ transformants (Fig. 3C). The ~700bp KanMX5′ and KanMX3′ fragments direct the mutagenized YFG library into the yfgΔ::KanMX genomic locus via homologous recombination, with retention of the original bar codes flanking each gene (Fig. 3C). Pools of Ura+ cells containing the mutant alleles are sporulated (Fig. 3E). Spores thus formed are spread on a haploid selective medium and incubated at 25°C for colony formation. Only haploid MATa Ura+ spores containing the integrated mutant allele of YFG can grow on this medium. Colonies formed on selective medium are replica-plated and incubated at 37°C (Fig. 3F). Colonies growing at 25°C but not at 37°C are selected as potential Ts alleles, and retested. In summary, the final product of the diploid shuffle approach is a confirmed MATa strain from the YKO collection genetic background containing a URA3 marked Ts allele of a specific gene integrated into its endogenous locus and flanked by both barcodes. In addition, each strain contains a LEU2-MFA1pr-HIS3 reporter integrated at the CAN1 locus.
Fig. 3. A diagram of the “Diploid shuffle” method for generating temperature sensitive alleles.
(A.) Genomic DNA containing YFG and its 5′ and 3′ regions is used as the template for PCR mutagenesis. Two black horizontal arrows represent the gene-specific primers used. The mutagenized PCR products are cloned into the vector SB221+Topo-TA (mutations are represented by black stars). The Topo-TA cloning site is represented by a black T, the A overhang protruding from the PCR product is represented by a black A. Left gray bar represents the 5′ half of the KanMX selectable marker (Kan), the right gray bar represents the other half of the KanMX selectable marker (MX). The NotI restriction sites are indicated by 2 diagonal black arrows. (B.) The product of the cloning step is a library of a mutagenized YFEG. The library is then transformed into E. coli, and digested with NotI to release linear fragments (following DNA purification). (C.) The linearized library is transformed into the corresponding heterozygous diploid strain. Bars that flank the KanMX knockout represent the two Barcodes. (D.) Heterozygous diploid transformants are sporulated (following meiosis), and MATa Ura+ haploids spores are selected on haploid selective medium at 25°C. (E.) Selection of temperature sensitive candidates following the replica plating and incubating at 25°C C and 37°C. Back arrows identify a potential Ts allele.
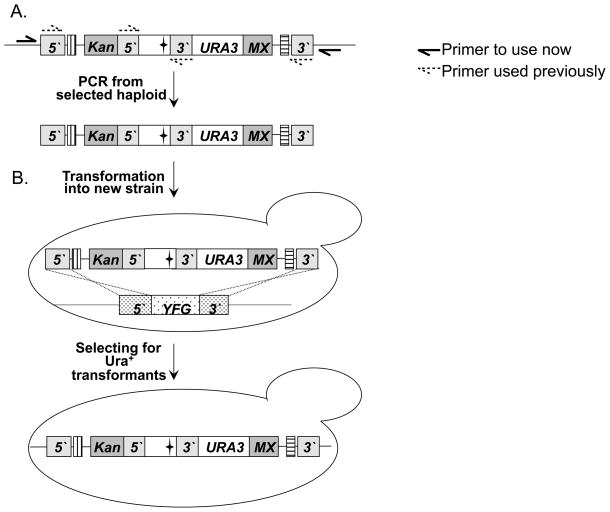
In addition to creating Ts alleles, the diploid shuffle-chromosomal method can also be used to transfer existing alleles into the knockout strain background from other strain backgrounds and vice versa. Using the Topo-TA plasmid and protocol described in Fig. 1, any extant Ts allele can be easily transferred to the deletion collection genetic background (referred to as “allele transfer-in”). The result is an integrated allele, marked by URA3, and flanked by the appropriate barcodes. Moreover, using the Topo-TA plasmid, any PCR product (mutant allele, fusion protein, heterologous gene expression cassette, etc.) can be introduced at any of the 6000 genomic sites carrying a KanMX replacement cassette as the integration site, depending on the specific deletion mutant chosen as the recipient strain. By using primers that are external to the primers used for the original mutagenesis, the URA3 marked Ts-allele or gene construct can be easily transferred from the deletion set genetic background to any other strain of interest, and replace the wild-type copy in the recipient strain by homologous recombination (referred to as “allele transfer-out”, see Fig. 4). Thus, each Ts mutation or gene construct can be analyzed for more specific phenotypes of interest in a variety of genetic contexts.
Fig. 4. Allele transfer-out.
Unless otherwise stated, all the symbols are as in Fig. 3 (A.) In this specific example, genomic DNA containing a Ts allele that was generated by the diploid shuffle method PCR amplified. Two black arrows represent the primers used for amplification. These primers are specific to the 5′ and 3′ regions of YFG in the recipient strain. They are also external to the two primers originally used to generate the Ts allele in the donor strain (represented by two broken arrows). (B.) The PCR product is transformed to the strain of interest. The specific example shows allele transfer to a haploid strain. However, it may be desirable to transfer the allele to a diploid strain first, followed by sporulation and tetrad dissection, if the Ts allele being transferred is inviable in the specific genetic context of interest. The Ts allele replaces the wild-type YFG by homologous recombination (represented by dashed lines) and give rise to Ura+ transformants (indicated by the Ts phenotype).
Materials
Media
Haploid selection medium SC–Ura–Leu–His–Arg+Can: dextrose, 20g/L; yeast nitrogen base without amino acids and ammonium sulfate, 1.7g/L; SC-Ura-Leu-His-Arg dropout mix, 2g/L; sodium glutamate, 1g/L; L-canavanine, 60mg/L; Agar, 2%. YPD: yeast extract, 10g/L; peptone, 20g/L; dextrose, 20g/L; Agar, 2%. YPD+G418: yeast extract, 10g/L; peptone, 20g/L; dextrose, 20g/L; Agar, 2%; G418, 200 mg/L.
Diploid selection medium SC-Ura+ClonNAT: dextrose, 20 g/L; yeast nitrogen base without amino acids and ammonium sulfate, 1.7 g/L; SC–Ura dropout mix, 2g/L; sodium glutamate, 1g/L; ClonNAT, 200 mg/L; Agar, 2%. The sodium glutamate is substituted for ammonium sulfate as the nitrogen source and makes the ClonNAT selection more reliable on the minimal medium.
Others are the same as described in Part I.
Strains
The genotype of haploid-convertible heterozygous diploid strains are similar to those described in Part I. Haploid Ts strains have the following genotype: MATa ura3Δ0 leu2Δ0 his3Δ1 lys2Δ0 (or LYS2) met15Δ0 (or MET15) can1Δ::LEU2-MFA1pr::HIS3 yfg-ts::URA3.
BY4742-ade2101-NatMX is a MATα wild-type haploid strain in which the NatMX gene is linked to the ade2-101 ochre mutation. This strain is used to confirm the MATa Ura+ Ts candidate: MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 ade2-101-NatMX
OneShot® TOP 10 Electrocomp Cells (Invitrogen Cat #C4040-52)
Plasmids
SB221 was derived from M4758 (Voth et al., 2003). In M4758, the BglII/KpnI and SphI/EcoRI fragments contain the TEF promoter (388bp) and terminator (262bp) respectively of Ashbya gossypii. In SB221 these fragments were replaced by a BamHI/KpnI PCR fragment (731bp) containing the TEF promoter plus half of the KanMX gene, and a SphI/EcoRI fragment (751bp) containing the other half of the KanMX gene and the TEF terminator. In both cases the template for PCR products was the KanMX gene used for constructing the heterozygous diploid collection. Finally the BamHI site of SB221, was adjusted with a Topo-TA site (invitrogen) to create SB221-Topo-TA.
Methods
PCR mutagenesis
This can be carried out as described in Part I. However, we have successfully used a slightly different condition for all the experiments using this diploid shuffle chromosomal method. Here we have exclusively used LA Taq DNA polymerase (Cat# RR002B) and 150 μM MnCl2. One PCR of 50 μl, instead of four, is normally set up for each gene. Two primers, which allow amplification of the entire coding region, 250–300bp of the 5′UTR, and 150–200bp of the 3′UTR of each gene, are used for PCR.
Cloning the PCR products
The mutagenized PCR products are purified and cloned into E. coli cells via electroporation.
Purify the PCR product using the ChargeSwitch® Clean-up Kit (Invitrogen Cat #CS12000).
Set up a ligation reaction that includes the following components: 0.5μl SB221 TOPO-TA, 1.0 μl ligation Buffer (300mM NaCl, 15mM MgCl2), purified PCR product (100ng/1kb), and ddH2O to a total volume of 6 μl.
Incubate the reaction at room temperature overnight.
Store the reaction at 4°C (if using soon) or at (−20°C).
Take 1.5 μl of the ligation products and transform into an aliquot of 50 μl OneShot® TOP 10 Electrocomp Cells (Invitrogen Cat #C4040-52) via electroporation.
Add 200 μl of LB into the transformation and incubate at 37°C for 1 hour.
Plate a 1 μl aliquot onto a LB plus ampicillin (LB-amp) plate to estimate the transformation efficiency.
Transfer 100 μl of the transformation suspension into a flask containing 50 ml of LB-amp liquid media and incubate at 37°C for ~12h to amplify the plasmid library. If necessary, store the rest of the transformation suspension (approximately 150 μl) in 20% glycerol at −70°C for later use.
Count the number of colonies on the 1 μl plate and multiply by the microliters inoculated into the LB-amp liquid media to estimate the complexity of the library. (The total number of colonies should be ~150,000–200,000)
Purify the plasmid DNA library using a Plasmid Midi prep kit (Qiagen Cat #12745).
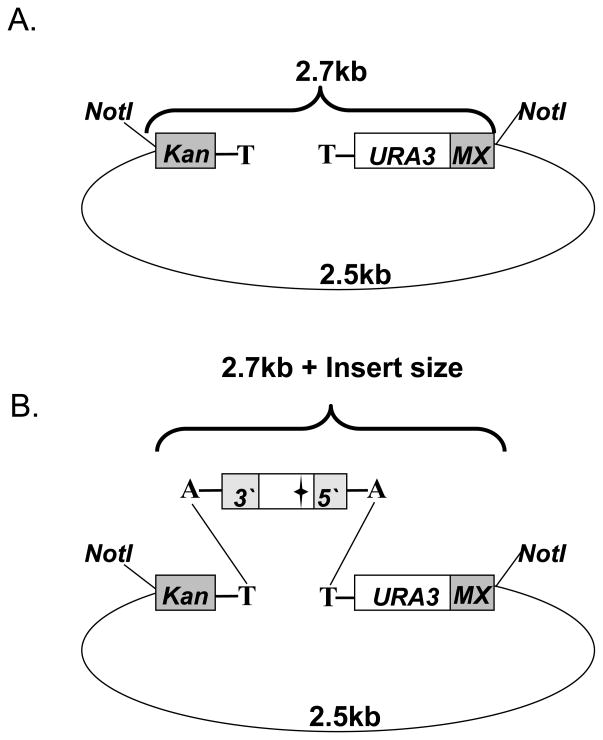
Digest 2 μl of 10x diluted library DNA sample with NotI (10U/μl, NEB, Cat#R018S) in a 10 μl reaction and examine the digests with agarose gel electrophoresis. This will allow estimation of the overall ligation efficiency. 2.5kb+2.7kb fragments represent the empty vector (Fig. 5A), while a successful cloning reaction is indicated by a band shift of the 2.7kb fragment (in accordance with the insert size) (Fig. 5B).
Fig. 5. Testing the Topo-TA cloning efficiency in SB221.
Unless otherwise stated, all the symbols are as in Fig. 3 (A.) The SB221 vector is made up of the URA3 gene flanked by a fragment containing the TEF promoter plus half of the KanMX gene (Kan), and another fragment containing the other half of the KanMX gene and the TEF terminator (Mx). This 2.7kb fragment can be excised from the 2.5kb backbone by restriction digestion with NotI. (B.) Restriction digest with NotI following a successful Topo-TA cloning reaction is indicated by the 2.5kb backbone and a band shift of the 2.7kb fragment, in accordance with the insert size.
Yeast transformation
Purified plasmid DNA sample of the random mutagenesis library is next digested with NotI or EcoRI (NEB, Cat#R0101T, 100u/μl) and transformed into the corresponding haploid-convertible heterozygous knockout diploid mutant. The mutant alleles will be integrated into the endogenous locus via homologous recombination.
Digest 40 μg of library plasmid DNA (over night at 37°C) in a total volume of 400ul using either of the two restriction enzymes mentioned above.
Reduce the reaction volume to 100 μl using a speed-vac.
Use the digested library DNA to transform the corresponding heterozygous diploid strain. Split the digested DNA to two 50 μl aliquots, and use each of them to prepare a separate transformation suspension as already described (Pan et al., 2004).
Combine the two transformation suspensions (~1284 μl) and plate 1 μl onto a SC–Ura plate to estimate the transformation yield.
Transfer the rest of the transformation suspension to 50 ml liquid SC–Ura and incubate at 30°C for 2 days to amplify the yeast library.
Count the number of colonies on the 1 μl plate and multiply by the microliters inoculated into the SC–Ura to estimate the total transformation yield. This number should be no less than 15,000. Otherwise, repeat yeast transformation.
Yeast sporulation
Amplified yeast library is then sporulated to convert the heterozygous diploid into haploid spores similarly as described in Part I. However, a slightly different haploid selection medium, SC–Ura–Leu–His–Arg+Can, is used to determine the efficiency of producing haploid MATa Ura+ cells from the sporulation culture.
Screening for Ts alleles
After the plating efficiency is determined, typically ~6000 haploid MATa Ura+ colonies are screened for candidate Ts alleles for each gene.
Spread the sporulation culture on 15 plates of SC–Ura–Leu–His–Arg+Can at ~400 colonies per plate.
Incubate these plates at 25°C for 3–4 days.
Replica-plate colonies on each plate to a fresh plate of the same haploid selection media and mark the orientation of both the mother and the daughter plates.
Incubate the mother plates at 25°C and the daughter plates at 37°C.
Assess the Ts phenotype of each clone by comparing its growth on both mother and daughter plates on the next day.
Confirming Ts phenotypes
Candidate Ts mutants are re-streaked on two haploid selection media plates, and incubate at 25°C and 37°C. Once the Ts phenotype is confirmed, backcross the MATa Ura+ Ts candidate to a wild-type BY4742-ade2101-NatMX MATα strain. In this strain the NatMX gene (which provides resistance to the drug ClonNAT) is linked to the ade2-101 ochre mutation. Diploid cells are selected by streaking on SC-Ura+ClonNAT media, sporulated, and dissected. The dissected tetrads are replicated to YPD (25°C and 37°C), SC-Ura (25°C), and YPD supplemented with G418 (25°C). This confirms that: 1) the temperature sensitivity segregates in a Mendelian manner (2:2), and indicates that the Ts phenotype depends on a single mutated gene, 2) the Ts phenotype is linked to the URA3 gene, and therefore co-segregates with the mutated PCR product, 3) the mutagenized PCR product was integrated at the correct genomic locus rendering the cells G418 sensitive (5′kanMX::yfg-ts-URA3::3′kanMX).
Allele transfer-in
This is carried out similarly as described above but with subtle modifications. A pre-existing Ts allele (or other gene construct) is first PCR-amplified with the appropriate plasmid or genomic DNA as the template using a proofreading-competent polymerase (e.g. LA Taq DNA polymerase) that generate an “A” overhang on each 3′ end of the PCR product. The PCR products are cloned into SB221 using Topo TA cloning. The cloned PCR products are then released together with the URA3 marker and the KanMX5′ and KanMX3′ fragments from the vector backbone and trans formed into the corresponding haploid convertible heterozygous diploid mutant. Ura+ yeast transformants are sporulated as a population and plated on SC–Ura–Leu–His–Arg+Can to select for haploid MATa Ura+ cells at an appropriate colony density. Single colonies are then tested for the phenotype of interest (e.g. temperature sensitivity). Candidate clones are then backcrossed to a wild-type MATα strain and analyzed with tetrad dissection to further confirm the phenotype.
Allele transfer-out
Here we describe how to transfer a Ts allele generated by the diploid shuffle method to any other ura3 strain background. Unless otherwise stated, methodologies are as mentioned above.
Design PCR primers that will amplify the whole 5′kanMX::yfg-ts-URA3::3′kanMX cassette. These primers are specific to the 5′ and 3′ regions of YFG in the recipient strain. They should also be external to the two primers originally used to generate the Ts allele in the donor strain (Fig. 4A).
Set up a PCR using genomic DNA of the Ts mutant as DNA template.
Transform the PCR product into your strain of interest and select for Ura+ transformants.
If the recipient strain is haploid, screen for colonies with a Ts phenotype.
Backcross to a wild-type strain of the opposite mating type.
Verify that the Ts phenotype is linked to the Ura+ by tetrad analysis.
The recipient strain can also be diploid. In this case, Ura+ transformants are selected after Step 3, sporulated, and characterized by tetrad analysis to ensure that the Ts and Ura+ phenotypes co-segregate. This will also allow testing whether the Ts allele is viable in the particular genetic context of the recipient strain. It is also possible that this allele is no longer Ts in this strain background. If so, representative Ura+ transformants will need to be characterized with diagnostic PCR or sequencing to ensure that the 5′kanMX::yfg-ts-URA3::3′kanMX cassette is indeed integrated at the right locus.
Perspectives
The two variations of the “diploid shuffle” are both highly efficient methods for making Ts mutants. It is essentially always possible, by screening enough mutants using the methods outlined here, to find such alleles. An adaptation of the methods outlined here will be required for making Ts mutants in very large essential genes (in this case, the gene would be mutagenized in sections). The relative advantages of the methods described here are summarized in Table 4.
We have chosen to move forward with the chromosomal method for the generation of a genome-wide collection of Ts mutants as a community resource because it was felt that most users would prefer integrated copies that would not fluctuate or be lost from a subpopulation of cells at each division due to their being on an episome.
The ability to generate a genome wide collection compares favorably with other attempts to generate genome wide resources for the study of essential genes, such as Tet-regulated alleles (Hartman et al., 2001; Mnaimneh et al., 2004) and dAMP (Schuldiner et al., 2005). Both of those approaches, while having their distinct advantages, only produced well-behaved alleles in about 30% of the cases. Disadvantages of Ts mutants include the fact they are not uniform with regard to nonpermissive temperature and leakiness, and the fact that the needed temperature shifts may induce heat shocks or other side effects that could potentially cloud phenotypic analyses. Nevertheless these alleles have been the bastion of traditional genetic analyses of essential genes. The Ts alleles we have sequenced include a mix of single amino acid substituions and multi amino acid substitutions; however, we have not sequenced enough of these to develop extensive statistics on this. Studies of collections of Ts mutants in a variety of genes have allowed the empirical determination of their typical characteristics. On this basis, a schemes for predicting Ts mutants has been developed (Ye, P., Dymond, J., Shi, X., Lin, Y.-Y., Pan, X., Boeke J.D. and Bader, J.S., submitted). This is likely to prove very useful for designing Ts mutants for use in other organisms in which extensive screening is impractical.
Table 4.
Advantages of the two methods.
| Plasmid | Chromosomal | Both |
|---|---|---|
| Faster/easier | Mutant is single copy | Works for any essential gene |
| No special reagents needed | Mutant is stable (not episomal) | Mutants are tagged with molecular barcodes assigned to original knockout |
References
- Ben-Aroya S, et al. Toward a Comprehensive Temperature-Sensitive Mutant Repository of the Essential Genes of Saccharomyces cerevisiae. Mol Cell. 2008;30:248–58. doi: 10.1016/j.molcel.2008.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann CB, et al. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–32. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Edgar RS, Lielausis I. Temperature-Sensitive Mutants of Bacteriophage T4d: Their Isolation and Genetic Characterization. Genetics. 1964;49:649–62. doi: 10.1093/genetics/49.4.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz RD, et al. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast. 1995;11:355–60. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- Gietz RD, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–34. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- Hartman JLt, et al. Principles for the buffering of genetic variation. Science. 2001;291:1001–4. doi: 10.1126/science.291.5506.1001. [DOI] [PubMed] [Google Scholar]
- Hartwell LH. Macromolecule synthesis in temperature-sensitive mutants of yeast. J Bacteriol. 1967;93:1662–70. doi: 10.1128/jb.93.5.1662-1670.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horowitz NH. Biochemical genetics of Neurospora. Adv Genet. 1950;3:33–71. doi: 10.1016/s0065-2660(08)60082-6. [DOI] [PubMed] [Google Scholar]
- Huang Z, et al. Plasmid-chromosome shuffling for non-deletion alleles in yeast. Nat Methods. 2008;5:167–9. doi: 10.1038/nmeth.1173. [DOI] [PubMed] [Google Scholar]
- Hughes TR. Yeast and drug discovery. Funct Integr Genomics. 2002;2:199–211. doi: 10.1007/s10142-002-0059-1. [DOI] [PubMed] [Google Scholar]
- Inoue H, et al. High efficiency transformation of Escherichia coli with plasmids. Gene. 1990;96:23–8. doi: 10.1016/0378-1119(90)90336-p. [DOI] [PubMed] [Google Scholar]
- Jeong H, et al. Lethality and centrality in protein networks. Nature. 2001;411:41–2. doi: 10.1038/35075138. [DOI] [PubMed] [Google Scholar]
- Li MZ, Elledge SJ. Harnessing homologous recombination in vitro to generate recombinant DNA via SLIC. Nat Methods. 2007;4:251–6. doi: 10.1038/nmeth1010. [DOI] [PubMed] [Google Scholar]
- Leung DW, Chen E, Goeddel DV. A method for random mutagenesis of a defined DNA segment using a modified polymerase chain reaction. J Cell Mol Biol. 1989;1:11–15. [Google Scholar]
- Lin YY, et al. A comprehensive synthetic genetic interaction network governing yeast histone acetylation and deacetylation. Genes Dev. 2008;22:2062–74. doi: 10.1101/gad.1679508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mnaimneh S, et al. Exploration of essential gene functions via titratable promoter alleles. Cell. 2004;118:31–44. doi: 10.1016/j.cell.2004.06.013. [DOI] [PubMed] [Google Scholar]
- Pan X, et al. A DNA integrity network in the yeast Saccharomyces cerevisiae. Cell. 2006;124:1069–81. doi: 10.1016/j.cell.2005.12.036. [DOI] [PubMed] [Google Scholar]
- Pan X, et al. A robust toolkit for functional profiling of the yeast genome. Mol Cell. 2004;16:487–96. doi: 10.1016/j.molcel.2004.09.035. [DOI] [PubMed] [Google Scholar]
- Schuldiner M, et al. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell. 2005;123:507–19. doi: 10.1016/j.cell.2005.08.031. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voth WP, et al. New ‘marker swap’ plasmids for converting selectable markers on budding yeast gene disruptions and plasmids. Yeast. 2003;20:985–93. doi: 10.1002/yea.1018. [DOI] [PubMed] [Google Scholar]