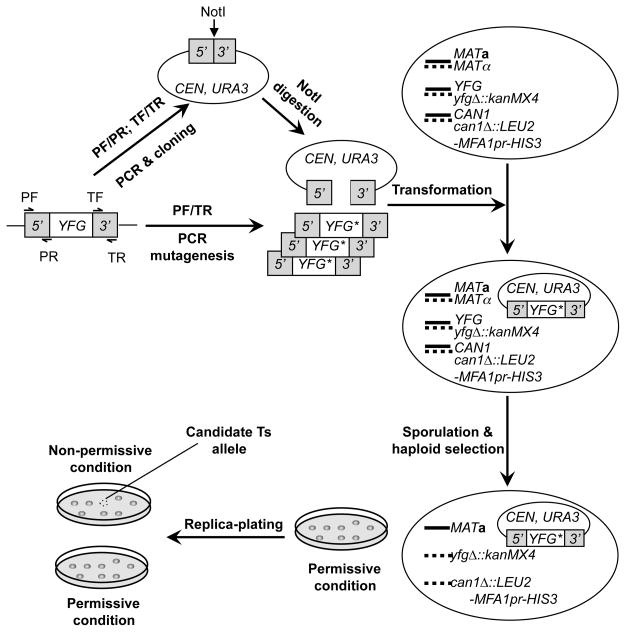
Fig. 1. A schematic for creating conditional alleles using plasmid-chromosome shuffle.
The promoter (5′) and terminator (3′) of YFG are separately PCR-amplified with primer pairs PF/PR and TF/TR, respectively, and cloned together onto a centromeric (CEN) yeast-E. coli shuttling vector. Here PF stands for promoter forward, PR for promoter reverse, TF for terminator forward, and TR for terminator reverse. A NotI recognition site is engineered between the promoter and terminator. The resultant promoter/terminator clone is linearized with NotI digestion. In the meanwhile, the entire sequence of YFG gene, including the coding region and the promoter and terminator sequences, is mutagenized using error-prone PCR with the primer pair PF/TR. The mutagenesis PCR products and the linearized promoter/terminator plasmid DNA are mixed and transformed together into a haploid-convertible heterozygous diploid knockout mutant of the same gene (MATa/α YFG/yfgΔ::kanMX4 CAN1/can1Δ::LEU2-MFA1pr-HIS3). The linearized promoter/terminator clones are repaired inside yeast cells mostly via homologous recombination using the co-transformed mutagenesis products (or YFG* alleles) as the templates. Due to the extensive homology between the ends of the PCR product and the vector, >105 recombinant clones can be easily generated. This pool of recombinants are then sporulated. Haploid MATa G418R Ura+ cells are selected under a permissive condition on solid SC-Ura-Leu-His-Arg+G418+Can medium as single colonies, which are replica-plated onto two fresh plates and incubated under both permissive and non-permissive conditions. Candidate alleles will grow under the permissive condition but not the non-permissive condition.