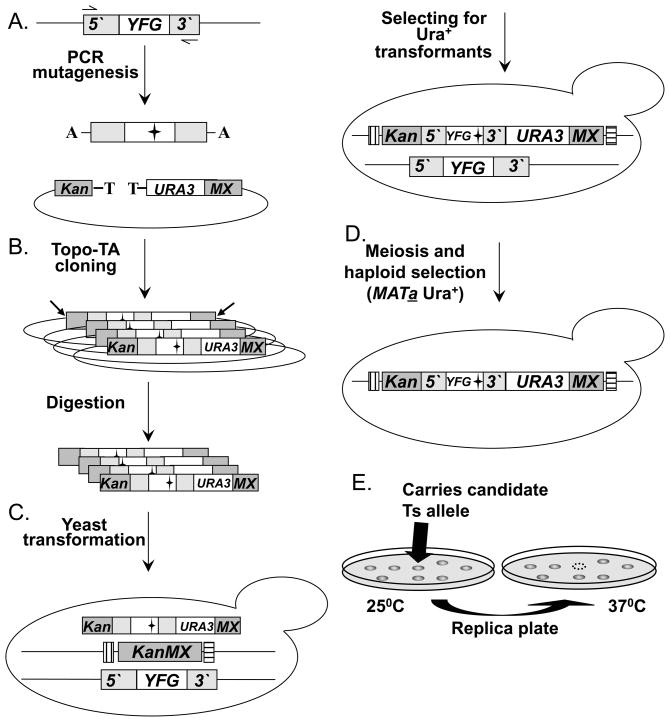
Fig. 3. A diagram of the “Diploid shuffle” method for generating temperature sensitive alleles.
(A.) Genomic DNA containing YFG and its 5′ and 3′ regions is used as the template for PCR mutagenesis. Two black horizontal arrows represent the gene-specific primers used. The mutagenized PCR products are cloned into the vector SB221+Topo-TA (mutations are represented by black stars). The Topo-TA cloning site is represented by a black T, the A overhang protruding from the PCR product is represented by a black A. Left gray bar represents the 5′ half of the KanMX selectable marker (Kan), the right gray bar represents the other half of the KanMX selectable marker (MX). The NotI restriction sites are indicated by 2 diagonal black arrows. (B.) The product of the cloning step is a library of a mutagenized YFEG. The library is then transformed into E. coli, and digested with NotI to release linear fragments (following DNA purification). (C.) The linearized library is transformed into the corresponding heterozygous diploid strain. Bars that flank the KanMX knockout represent the two Barcodes. (D.) Heterozygous diploid transformants are sporulated (following meiosis), and MATa Ura+ haploids spores are selected on haploid selective medium at 25°C. (E.) Selection of temperature sensitive candidates following the replica plating and incubating at 25°C C and 37°C. Back arrows identify a potential Ts allele.