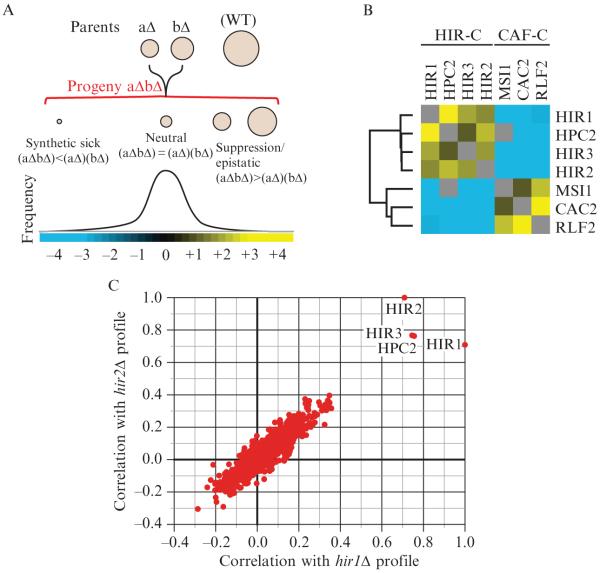
Figure 9.1.
Epistatic interactions within and between chromatin assembly complexes. (A) The entire spectrum of genetic interactions. Quantitative genetic analysis can identify negative ((aΔbΔ) < (aΔ)(bΔ)), positive ((aΔbΔ) > (aΔ)(bΔ)), and neutral ((aΔbΔ) = (aΔ)(bΔ)) genetic interactions. (B) Genetic interactions between and within the HIR and CAF chromatin assembly complexes. Using the E-MAP approach (Collins et al., 2007a), strong negative interactions were detected between components of the HIR-C (HIR1, HPC2, HIR3, and HIR2) and the CAF-C (MSI1, CAC2, and RLF2), which are known to function in parallel pathways to ensure efficient chromatin assembly. Conversely, positive genetic interactions were observed between components within each complex. Blue and yellow interactions correspond to negative and positive genetic interactions, respectively. (C) Plot of correlation coefficients generated from comparison of the genetic profiles from hir1Δ and hir2Δ to all other ~750 profiles from the chromosome biology E-MAP (Collins et al., 2007a). Note the high pairwise correlations with HIR1, HIR2, HIR3, and HPC2.