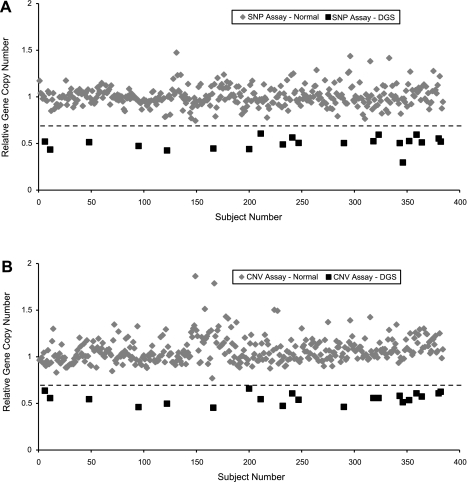
Fig. 1.
Scatter plots showing multiplexed quantitative real-time PCR (MQPCR) analyses over the TBX1 gene locus on human chromosome 22 for n = 382 subjects with congenital heart disease. A: single nucleotide polymorphism (SNP) assay. B: copy number variant (CNV) assay. A calculated relative gene copy number (rGCN) of 1.0 indicates a nondeleted subject (2 copies), whereas a rGCN of 0.5 indicates a hemizygous deletion (1 copy). Twenty-one subjects, denoted by squares, were labeled as deleted. The remaining 361 subjects, denoted by diamonds, were nondeleted. The dashed line represents a threshold value of 0.7, where the sensitivity and specificity of both assays is 100%. DGS, DiGeorge syndrome.