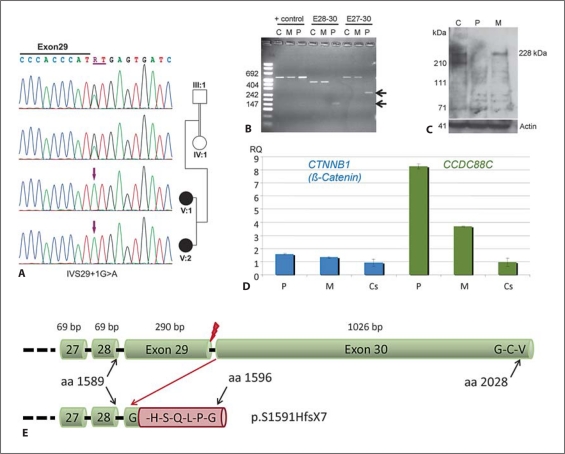
Fig. 4.
Characterization of the detected splice site mutation IVS29 + 1G>A. C = Control, M = Mother, P = Patient. A Electropherograms of the homozygous mutation detected in both affected siblings and heterozygous carrier status of both parents. B Confirmation of an aberrant transcript by reverse transcription PCR. The aberrant splicing in the patient caused by the mutation in intron 29 results in deletion of exon 29, which was confirmed by sequencing of the aberrant transcript. Deletion of exon 29 (290 bp) results in a frameshift in the last exon and subsequent loss of the terminal Dishevelled binding site. C Western blotting using an antibody against the epitope encoded by exons 28 and 29 confirmed absence of wild-type protein in the patient. D Results of quantitative RT-PCR using minor grove binding probes for CTNNB1 (β-catenin) and CCDC88C on cDNA derived from PAXgene RNA tube samples from the patient and her mother in comparison to 8 unrelated healthy controls. In line with the assumed loss of function without nonsense-mediated mRNA decay, we found significantly increased expression of both CCDC88C and β-catenin in the patient. The patient's heterozygous mother showed also increased CCDC88C expression levels but no statistical significant alteration of β-catenin expression (p < 0.028 in the patient, p < 0.148 in the mother; Wilcoxon two-sample test). E Scheme of the effect of the splice site mutation on the protein level. Upper cartoon showing the terminal part of the wild-type protein ending with the G-C-V Dishevelled binding site. Lower cartoon indicates the frameshift resulting from loss of exon 29 after the first amino acid from exon 30, the addition of 6 amino acids and the loss of the Dishevelled binding site (p.S1591HfsX7). Red flash indicates site of splice site mutation.